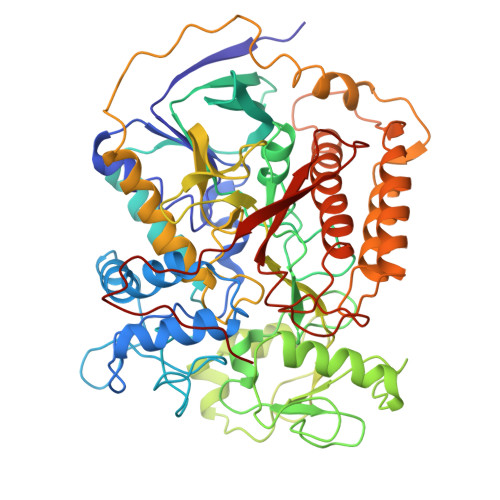
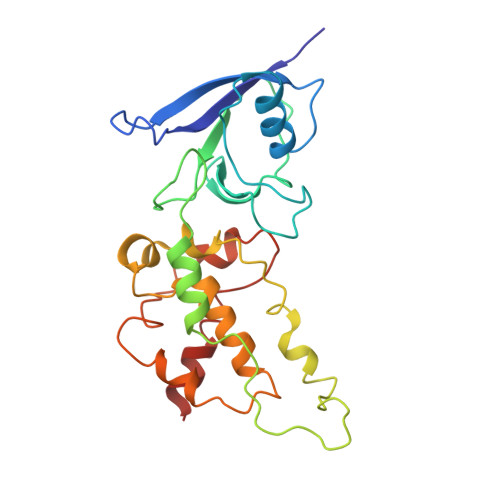
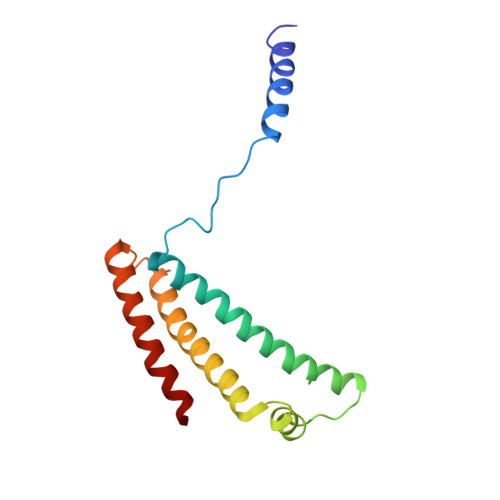
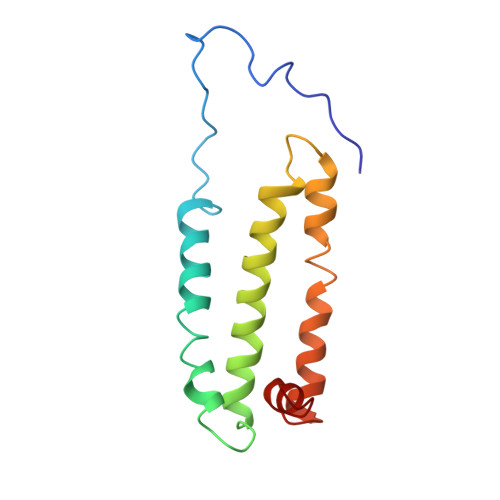
Cryo-EM structure of the yeast Saccharomyces cerevisiae SDH provides a template for eco-friendly fungicide discovery.
Li, Z.W., Huang, Y.H., Wei, G., Lu, Z.W., Wang, Y.X., Cui, G.R., Wang, J.Y., Yu, X.H., Fu, Y.X., Fan, E.D., Wu, Q.Y., Zhu, X.L., Ye, Y., Yang, G.F.(2025) Nat Commun 16: 8936-8936
- PubMed: 41062466
- DOI: https://doi.org/10.1038/s41467-025-64001-0
- Primary Citation of Related Structures:
9KPS, 9KPT, 9KQ3, 9LIG - PubMed Abstract:
Succinate dehydrogenase (SDH) is a key fungicidal target, but rational inhibitors design has been impeded by the lack of fungal SDH structure. Here, we show the cryo-EM structure of SDH from Saccharomyces cerevisiae (ScSDH) in apo (3.36 Å) and ubiquinone-1-bound (3.25 Å) states, revealing subunits architecture and quinone-binding sites (Q p ). ScSDH is classified as a heme-deficient type-D SDH, utilizing conserved redox centers (FAD, [2Fe-2S], [4Fe-4S] and [3Fe-4S] clusters) for electron transfer. A 3.23 Å structure with pydiflumetofen (PYD) identified critical interactions, including hydrogen bonds with Trp_SDHB194 and Tyr_SDHD120, and a cation-π interaction with Arg_SDHC97. Leveraging this, we designed a SDH inhibitor E8 (enprocymid), exhibiting significant fungicidal activity (K i = 0.019 μM) and reduced zebrafish toxicity (LC 50 (96 h) = 1.01 mg a.i./L). This study elucidates the structure of fungal SDH and demonstrates the potential of ScSDH for rational design of next-generation fungicides, addressing fungal resistance and environmental toxicity in agriculture.
- State Key Laboratory of Green Pesticide, International Joint Research Center for Intelligent Biosensor Technology and Health, Central China Normal University, Wuhan, P.R. China.
Organizational Affiliation: