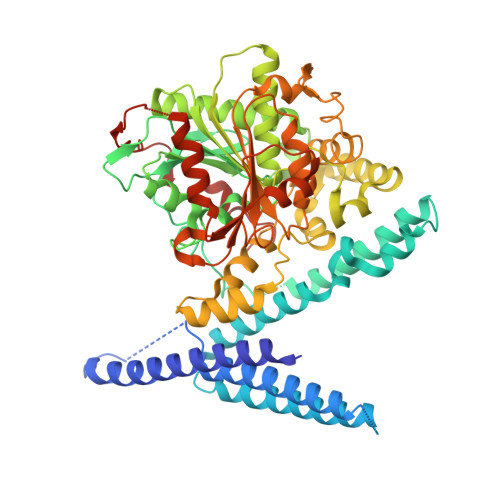
Structures of Polyhydroxyalkanoate Synthase PhaC from Aeromonas caviae, Producing Biodegradable Plastics.
Chek, M.F., Kim, S.Y., Mori, T., Matsumoto, K., Sato, S., Hakoshima, T.(2025) Angew Chem Int Ed Engl 64: e202504626-e202504626
- PubMed: 40276819
- DOI: https://doi.org/10.1002/anie.202504626
- Primary Citation of Related Structures:
9KNJ, 9KNK, 9KNL - PubMed Abstract:
Polyhydroxyalkanoate (PHA) is a biodegradable polyester that can serve as a promising alternative to petrochemical plastics, which present a serious source of pollution. PHA synthase (PhaC) is a key enzyme responsible for producing a wide variety of PHAs in microorganisms. Here, we present crystal structures of full-length PhaC from Aeromonas caviae, a high-performance PhaC employed for industrial use. The structure reveals an N-terminal helical domain that mediates head-to-head dimerization and stabilizes the C-terminal α/β catalytic domain to form a tunnel that connects the catalytic center embedded inside the protein to the protein surface. We showed that this tunnel is a putative egress tunnel for the product PHA chain. Our results establish a fundamental understanding of the PhaC machinery that should lead to improvement of this enzyme in industrial applications.
- Structural Biology and Protein Engineering Laboratory, Institute for Research Initiatives, Nara Institute of Science and Technology, 8916-5 Takayama, Ikoma, Nara, 630-0192, Japan.
Organizational Affiliation: