From Genome to structure: Elucidating Neurosporene Production in the Rhodobacter sphaeroides G1C Mutant
Wu, Y.-L., Yu, L.-J.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Reaction center protein L chain | A [auth L] | 282 | Cereibacter sphaeroides | Mutation(s): 0 | 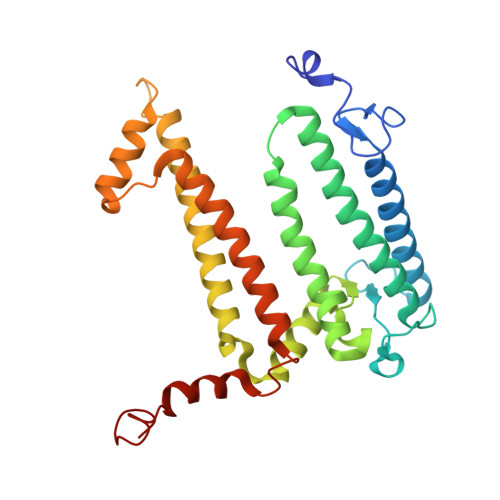 |
UniProt | |||||
Find proteins for P0C0Y8 (Cereibacter sphaeroides) Explore P0C0Y8 Go to UniProtKB: P0C0Y8 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0C0Y8 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Reaction center protein M chain | B [auth M] | 308 | Cereibacter sphaeroides | Mutation(s): 0 | 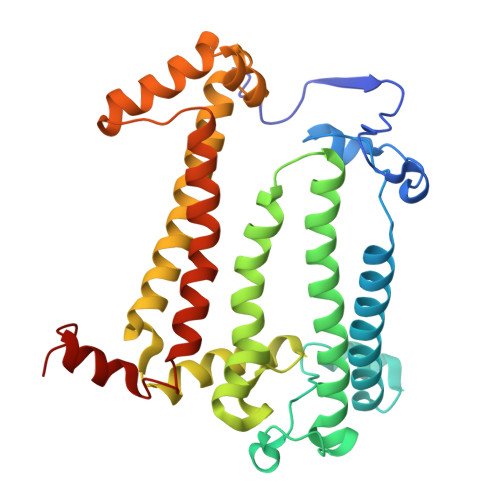 |
UniProt | |||||
Find proteins for P0C0Y9 (Cereibacter sphaeroides) Explore P0C0Y9 Go to UniProtKB: P0C0Y9 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0C0Y9 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Reaction center protein H chain | C [auth H] | 260 | Cereibacter sphaeroides | Mutation(s): 0 | 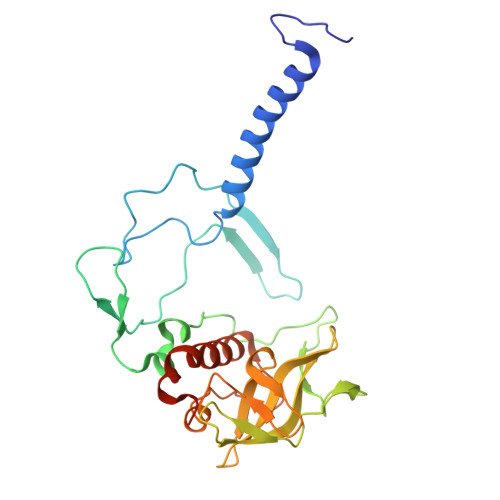 |
UniProt | |||||
Find proteins for P0C0Y7 (Cereibacter sphaeroides) Explore P0C0Y7 Go to UniProtKB: P0C0Y7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0C0Y7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Light-harvesting protein B-875 alpha chain | 54 | Cereibacter sphaeroides | Mutation(s): 0 | 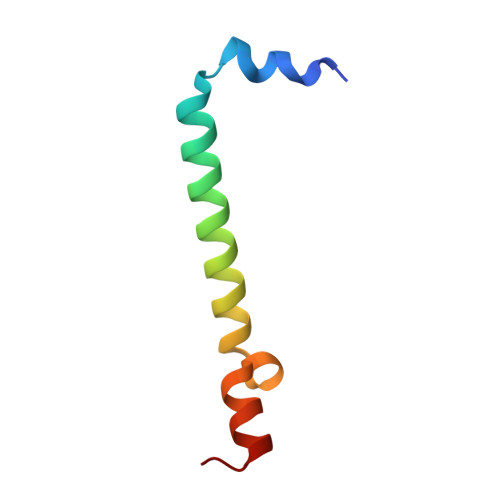 | |
UniProt | |||||
Find proteins for P0C0X9 (Cereibacter sphaeroides) Explore P0C0X9 Go to UniProtKB: P0C0X9 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0C0X9 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Antenna pigment protein beta chain | 49 | Cereibacter sphaeroides | Mutation(s): 0 | 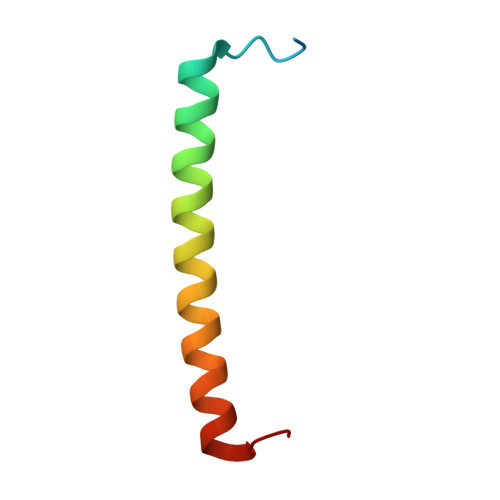 | |
UniProt | |||||
Find proteins for Q7B300 (Cereibacter sphaeroides) Explore Q7B300 Go to UniProtKB: Q7B300 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q7B300 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 6 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| protein-U | FA [auth U] | 53 | Cereibacter sphaeroides | Mutation(s): 0 | 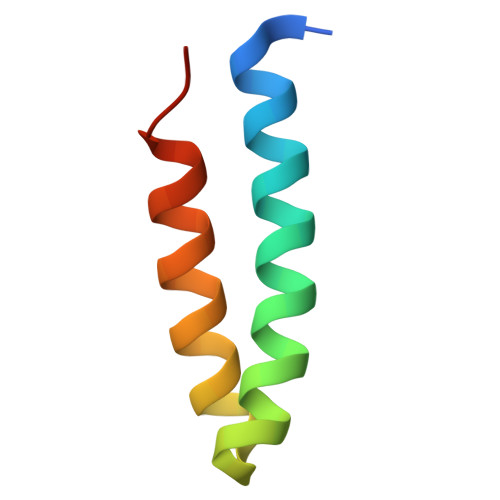 |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 7 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Intrinsic membrane protein PufX | GA [auth X] | 82 | Cereibacter sphaeroides | Mutation(s): 0 | 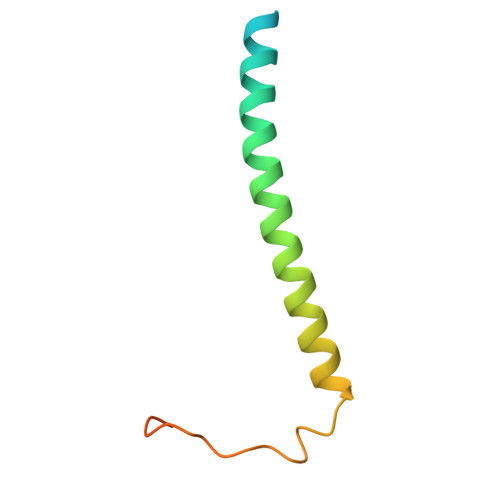 |
UniProt | |||||
Find proteins for Q7B2Z6 (Cereibacter sphaeroides) Explore Q7B2Z6 Go to UniProtKB: Q7B2Z6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q7B2Z6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 9 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| CDL (Subject of Investigation/LOI) Query on CDL | FB [auth M], IB [auth M], NC [auth E], VC [auth F] | CARDIOLIPIN C81 H156 O17 P2 XVTUQDWPJJBEHJ-KZCWQMDCSA-L |  | ||
| BCL (Subject of Investigation/LOI) Query on BCL | AC [auth C] DC [auth c] DD [auth G] DE [auth n] FD [auth g] | BACTERIOCHLOROPHYLL A C55 H74 Mg N4 O6 DSJXIQQMORJERS-AGGZHOMASA-M |  | ||
| BPH (Subject of Investigation/LOI) Query on BPH | AB [auth M], IA [auth L] | BACTERIOPHEOPHYTIN A C55 H76 N4 O6 KWOZSBGNAHVCKG-SZQBJALDSA-N |  | ||
| U10 (Subject of Investigation/LOI) Query on U10 | CB [auth M], JA [auth L], KA [auth L] | UBIQUINONE-10 C59 H90 O4 ACTIUHUUMQJHFO-UPTCCGCDSA-N |  | ||
| PGV (Subject of Investigation/LOI) Query on PGV | CD [auth G] HB [auth M] LA [auth L] LB [auth H] MA [auth L] | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE C40 H77 O10 P ADYWCMPUNIVOEA-GPJPVTGXSA-N |  | ||
| A1EF2 (Subject of Investigation/LOI) Query on A1EF2 | AE [auth N] BC [auth C] BE [auth N] CE [auth n] ED [auth g] | (6~{E},8~{E},10~{E},12~{E},14~{E},16~{E},18~{E},20~{E},22~{E},26~{E})-2,6,10,14,19,23,27,31-octamethyldotriaconta-2,6,8,10,12,14,16,18,20,22,26,30-dodecaene C40 H58 ATCICVFRSJQYDV-XILUKMICSA-N |  | ||
| LMT (Subject of Investigation/LOI) Query on LMT | AD [auth G] BD [auth G] CC [auth c] DB [auth M] EB [auth M] | DODECYL-BETA-D-MALTOSIDE C24 H46 O11 NLEBIOOXCVAHBD-QKMCSOCLSA-N |  | ||
| LDA (Subject of Investigation/LOI) Query on LDA | KD [auth I] NA [auth L] PA [auth L] QA [auth L] RD [auth J] | LAURYL DIMETHYLAMINE-N-OXIDE C14 H31 N O SYELZBGXAIXKHU-UHFFFAOYSA-N |  | ||
| FE (Subject of Investigation/LOI) Query on FE | BB [auth M] | FE (III) ION Fe VTLYFUHAOXGGBS-UHFFFAOYSA-N |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| FME Query on FME | A [auth L] | L-PEPTIDE LINKING | C6 H11 N O3 S |  | MET |
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | 1.19.2_4158: |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Ministry of Science and Technology (MoST, China) | China | 2022YFC3401800 |