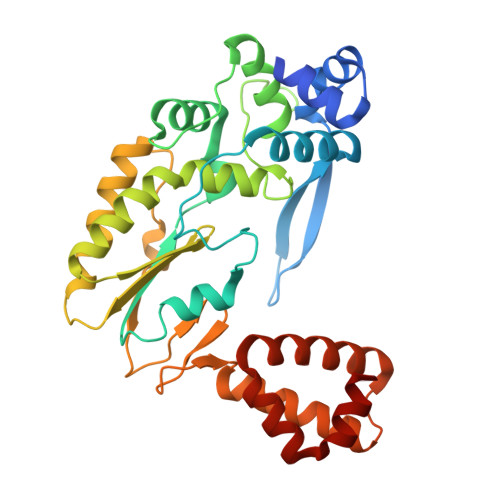
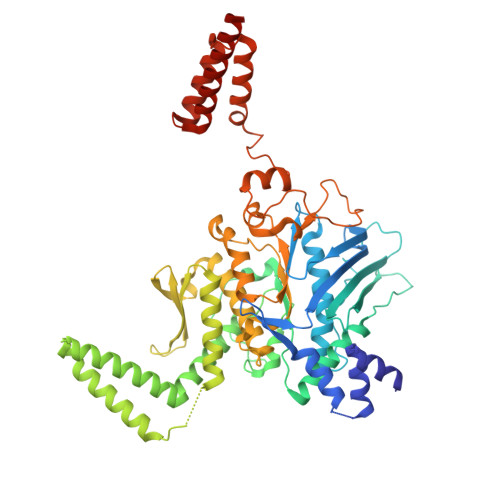
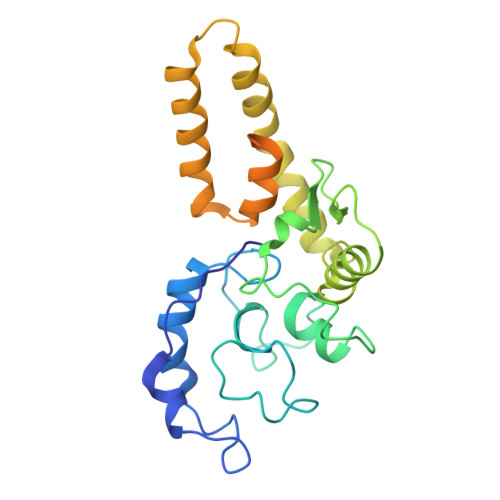
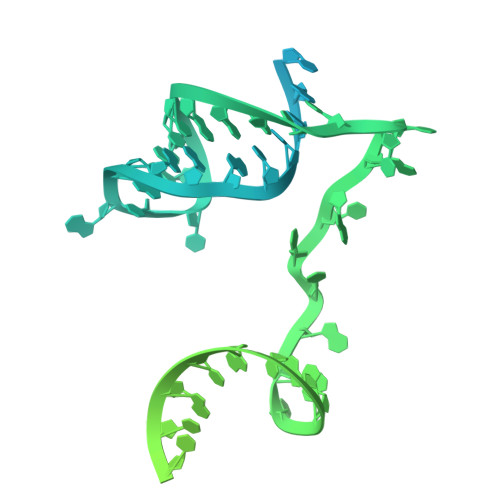
Structural mechanism of the Retron-Eco7 anti-phage defense system.
Ishikawa, J., Yoneyama, K., Azam, A.H., Nagao, A., Mitsuda, Y., Nakazaki, R., Chihara, K., Hiraizumi, M., Yamashita, K., Suzuki, T., Kiga, K., Nishimasu, H.(2025) Nat Commun 16: 10821-10821
- PubMed: 41330933
- DOI: https://doi.org/10.1038/s41467-025-66589-9
- Primary Citation of Related Structures:
9KJX, 9KJY, 9KJZ, 9KK1, 9KK2 - PubMed Abstract:
Retrons are prokaryotic genetic elements involved in anti-phage defense and consist of a non-coding RNA, a reverse transcriptase (RT), and various effector proteins. Retron-Eco7 (previously known as Retron-Ec78) from Escherichia coli encodes two effector proteins (the PtuA ATPase and the PtuB nuclease) and degrades the host tRNA Tyr upon phage infection, thereby protecting host cells against invading phages. However, its defense mechanism remains elusive. Here, we report the cryo-electron microscopy (cryo-EM) structures of the Retron-Eco7 complex, comprising the RT, multicopy single-stranded DNA (msDNA), PtuA, and PtuB. The Retron-Eco7 structures reveal that the RT-msDNA complex associates with two PtuA-PtuB complexes, potentially inhibiting their nuclease activity and suppressing bacterial growth arrest prior to phage infection. Furthermore, the phage-encoded D15 nuclease acts as a trigger for the Retron-Eco7 system and cleaves the msDNA bound to the complex, facilitating the dissociation of PtuA-PtuB from RT-msDNA. Our data indicate that msDNA cleavage by D15 is the initial step required for the specific cleavage of the host tRNA Tyr by the PtuA-PtuB nuclease, which leads to abortive infection. Overall, this study provides mechanistic insights into the Retron-Eco7 system and highlights the diversity of prokaryotic anti-phage defense mechanisms.
- Department of Chemistry and Biotechnology, Graduate School of Engineering, The University of Tokyo, Tokyo, Japan.
Organizational Affiliation: