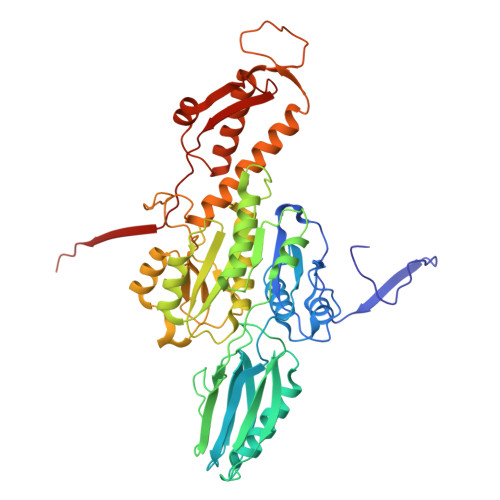
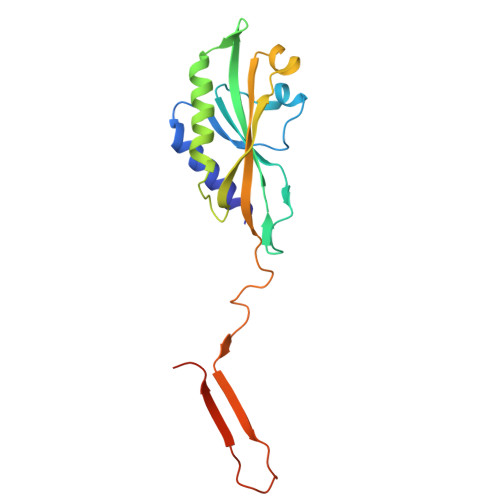
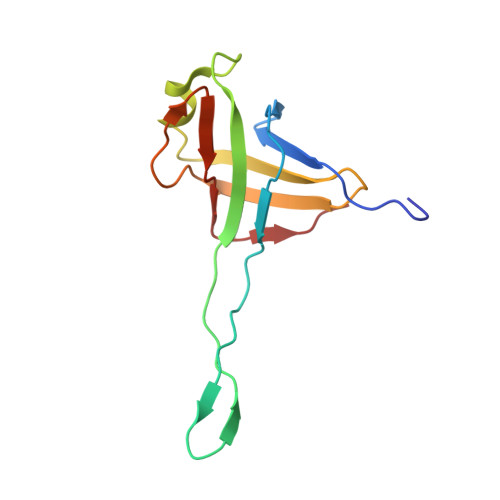
In situ structures of the contractile nanomachine myophage Mu in both its extended and contracted states.
Zhou, J., Wang, L., Xiao, H., Chen, W., Liu, Z., Song, J., Zheng, J., Liu, H.(2025) J Virol 99: e0205624-e0205624
- PubMed: 39992138
- DOI: https://doi.org/10.1128/jvi.02056-24
- Primary Citation of Related Structures:
9JOD, 9KHX, 9KHY, 9KI1, 9KNU, 9LJ8 - PubMed Abstract:
Myophage Mu is a representative of contractile nanomachines with a simple tail baseplate. It has the capacity to infect a range of intestinal bacteria and has extensive applications in genetic engineering research. Nevertheless, a comprehensive understanding of the entire structure and contractile mechanisms of Mu remains elusive. Using cryo-electron microscopy (cryo-EM), we resolved the asymmetric structures of Mu in both its extended and contracted states, the latter of which lacked the tail baseplate, at near-atomic resolutions. We built the atomic models for the extended Mu, encompassing the head, the connector complex, the tail, and the simple baseplate. It is noteworthy that we identified the position and structure of the tail tube initiator protein gp43 (referred to as the DNA circularization protein). The protein gp43 plays a crucial role not only in the baseplate assembly and DNA circularization but also in stabilizing the wedge-hub connection and mediating tail contraction. Except for the baseplate structure, the structural comparison of Mu in its extended and contracted states revealed that only the tail sheath protein gp39 and the C-terminus of the tail terminator protein gp37 undergo notable conformational changes to accommodate the tail contraction, whereas the remaining protein components remained unchanged. Our structures exhibited conserved properties among the majority of myophages, thereby providing valuable insights into the contraction mechanisms across myophages and contractile injection systems (CISs). Despite extensive study, the asymmetric structures of phage Mu, a highly effective transposable myophage, remain unknown. In this study, we present the high-resolution structures of Mu in both its extended and contracted states. The comparison of the two structures allows for the illustration of detailed conformational changes of the head-to-tail complex during the process of tail contraction. The contraction mechanism of Mu is highly conserved and widely adapted to all contractile nanomachines that share common structural features with Mu.
- Institute of Interdisciplinary Studies, Key Laboratory for Matter Microstructure and Function of Hunan Province, Key Laboratory of Low-Dimensional Quantum Structures and Quantum Control, School of Physics and Electronics, Hunan Normal University, Changsha, China.
Organizational Affiliation: