Structural Insights into the Force-Transducing Mechanism of a Motor-Stator Complex Important for Bacterial Outer Membrane Lipid Homeostasis.
Yeow, J., Chia, C.G., Lim, N.Z., Zhao, X., Yan, J., Chng, S.S.(2025) J Am Chem Soc 147: 24299-24308
- PubMed: 40589080
- DOI: https://doi.org/10.1021/jacs.4c18050
- Primary Citation of Related Structures:
9K49, 9KCH - PubMed Abstract:
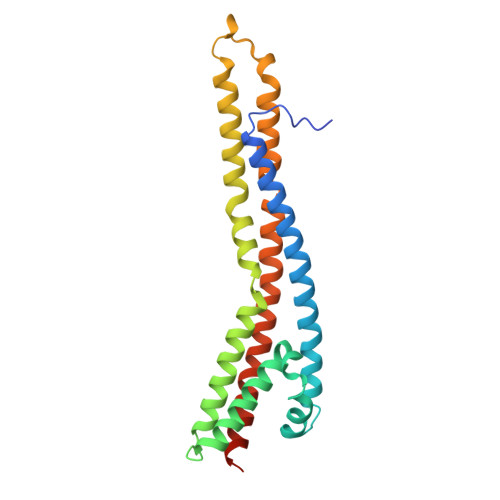
Gram-negative bacteria assemble an asymmetric outer membrane (OM) that functions as an effective barrier against antibiotics. Building a stable and functional OM requires the assembly and maintenance of balanced levels of proteins, lipopolysaccharides, and phospholipids into the bilayer. In Escherichia coli , the trans-envelope Tol-Pal complex has recently been established to play a primary role in maintaining OM lipid homeostasis. It is believed that the motor-stator complex TolQR exploits the proton motive force in the inner membrane to induce conformational changes in the TolA effector, ultimately generating a force across the cell envelope to activate processes at the OM. Molecular details of how such force transduction occurs via the TolQRA complex are unknown. Here, we solve structures of the E. coli TolQRA complex using single-particle cryo-EM, capturing the transmembrane (TM) regions of the purified complex in two distinct states at ∼3.6 and ∼4.2 Å nominal resolutions. We define how the TolA N-terminal TM helix interacts with an asymmetric TolQ 5 R 2 subcomplex in two different positions, revealing how the two TolQRA states are related by rotation of the TolQ pentamer. By considering structural prediction and biochemical evidence for the periplasmic domains of the complex, we propose a working model for how proton passage through the complex induces rotary movement that can be coupled to TolA for force transduction across the cell envelope.
- Department of Chemistry, Faculty of Science, National University of Singapore, 117543 Singapore.
Organizational Affiliation: