Structural insights into human signal peptide peptidase
Zhou, R., Huang, G., Guo, X., Wang, J., Ge, X., Shi, Y.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Signal peptide peptidase-like 2A | 520 | Homo sapiens | Mutation(s): 0 Gene Names: SPPL2A, IMP3, PSL2, PSEC0147 EC: 3.4.23 | 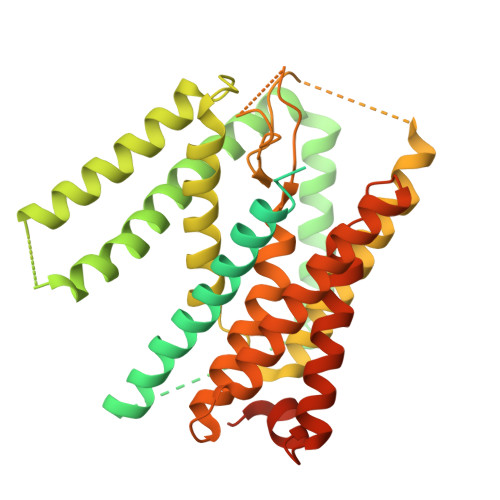 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q8TCT8 (Homo sapiens) Explore Q8TCT8 Go to UniProtKB: Q8TCT8 | |||||
PHAROS: Q8TCT8 GTEx: ENSG00000138600 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q8TCT8 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| PC1 (Subject of Investigation/LOI) Query on PC1 | B [auth A] | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE C44 H88 N O8 P NRJAVPSFFCBXDT-HUESYALOSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | |
| RECONSTRUCTION | cryoSPARC |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Basic Research Program of China (973 Program) | China | 2020YFA0509300 |