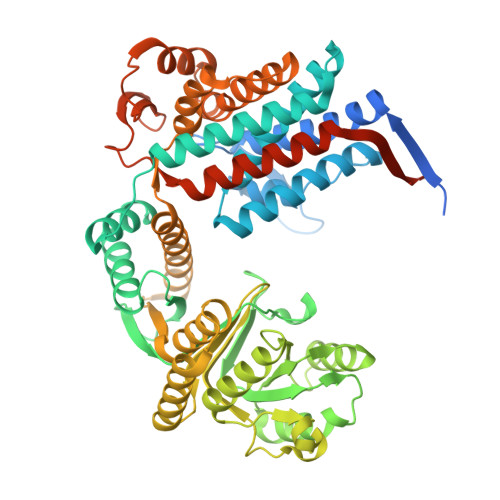
Soil aggregates stability is evidently enhanced by super-binding of the N-terminal disordered tail of glomalin to soil minerals
Jia, M., Wang, Y., He, J., Chou, S.H., Luo, D., Fang, L., Hou, J., Fan, Y., Zhang, W., Chen, H., Zhou, D., Liu, Y., Feng, Y., Koopal, L., Tan, W.(2025) Soil Biol Biochem 210: 109908