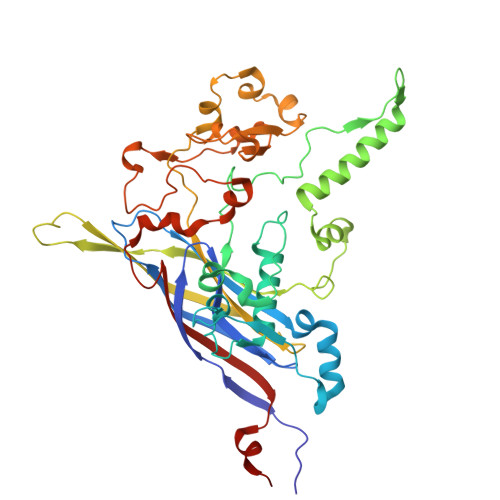
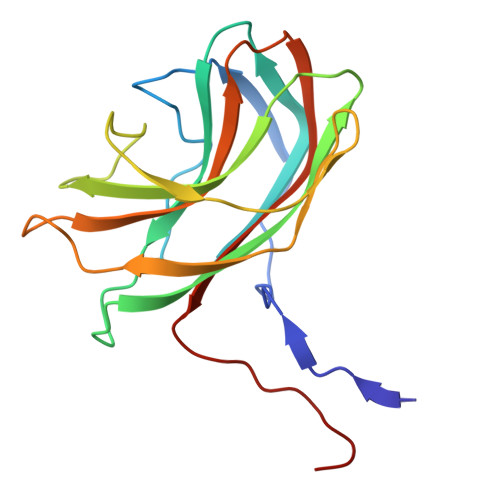
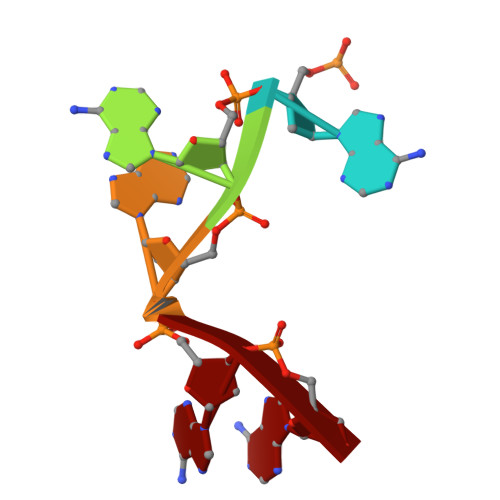
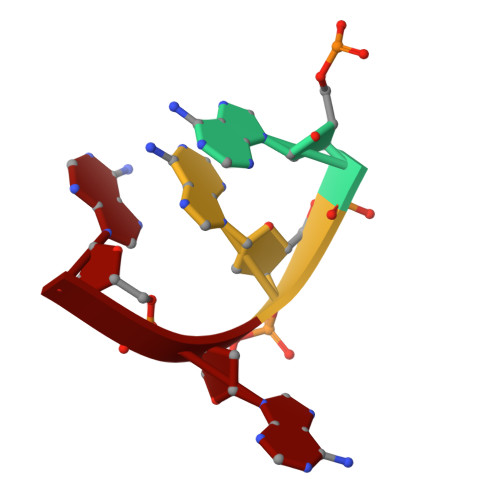
Structural basis for Salmonella infection by two Microviridae phages.
Hu, W., Liu, Z., Wei, Y., Bian, Q., Lan, W., Fan, C., Song, J., Sun, Q., Zhang, X., Liu, Y., Gao, Y., Chen, Y.(2025) Commun Biol 8: 1166-1166
- PubMed: 40770068
- DOI: https://doi.org/10.1038/s42003-025-08595-7
- Primary Citation of Related Structures:
9K3M, 9K3N - PubMed Abstract:
The global resurgence of multidrug-resistant Salmonella species, responsible for millions of annual infections, underscores the urgent need for alternative antimicrobial strategies, such as phage therapy. Microviridae phages offer a promising model for studying phage-host interactions with their unique structural and infection mechanisms. Here, we identify two Microviridae phages, PJNS001 and PJNS002, with different host receptor dependencies, and determine their cryo-EM structures at 2.68 Å and 2.59 Å resolution, respectively. These icosahedral capsids with T = 1 symmetry exhibit a unique vertex reinforcement mechanism, stabilizing the viral assembly. The specific pentameric adaptations, coupled with DNA binding protein engagements and thermodynamic constraints, collectively preclude the formation of hybrid virions. Structural analysis and in situ visualization reveal spike protein features and host-attachment intermediates, informing host specificity. Together, these findings advance our understanding of Microviridae infection mechanisms and provide a structural framework for rational phage design against antibiotic-resistant pathogens.
- Shanghai Institute for Advanced Immunochemical Studies, ShanghaiTech University, Shanghai, 201210, China.
Organizational Affiliation: