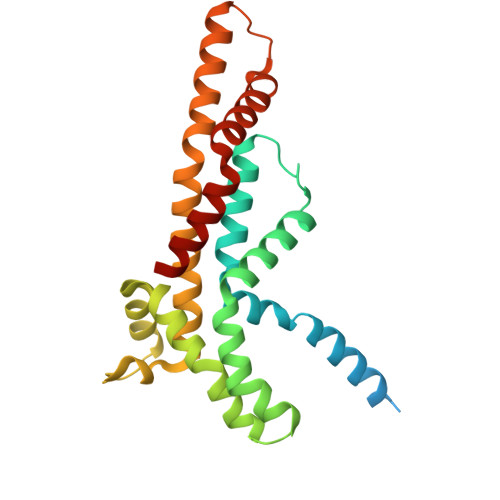
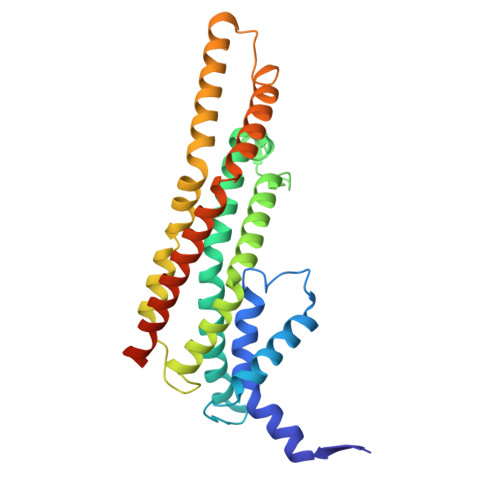
A beta-cap on the FliPQR protein-export channel acts as the cap for initial flagellar rod assembly.
Kinoshita, M., Miyata, T., Makino, F., Imada, K., Namba, K., Minamino, T.(2025) Proc Natl Acad Sci U S A 122: e2507221122-e2507221122
- PubMed: 40833400
- DOI: https://doi.org/10.1073/pnas.2507221122
- Primary Citation of Related Structures:
9K29 - PubMed Abstract:
The FliPQR complex constitutes a channel for export of the flagellar proteins involved in axial structure assembly. It also serves as a template for the assembly of the rod structure, which consists of FliE, FlgB, FlgC, FlgF, and FlgG. FliP, FliQ, and FliR assemble into a right-handed helical structure within the central pore of the flagellar basal body MS-ring, and the complex has two gates on the cytoplasmic and periplasmic sides. The periplasmic gate, formed by the N-terminal α-helices of FliP and FliR, remains closed until six FliE subunits assemble onto FliP and FliR to form the first layer of the rod, but it has remained unclear how each FliE subunit opens the gate and assembles in the absence of the rod cap required for efficient assembly of other rod proteins. Here, we present a cryoelectron microscopy structure of the FliPQR complex in closed form at 3.0 Å resolution. A β-cap, formed by the N-terminal β-strands of FliP and FliR, is located at the top of the FliPQR complex and tightly seals the closed gate. The β-cap has a narrow pore that efficiently and accurately leads the first FliE subunit to its assembly site. Interactions of FliE with FliP and FliR induce a conformational change in FliP and FliR, with their N-terminal α-helices move up and outward to open the gate. Consequently, each of the N-terminal β-strands of FliP and FliR detaches from the β-cap one after another, thereby creating a docking site for the next FliE subunit to efficiently assemble.
- Graduate School of Frontier Biosciences, Osaka University, Suita, Osaka 565-0871, Japan.
Organizational Affiliation: