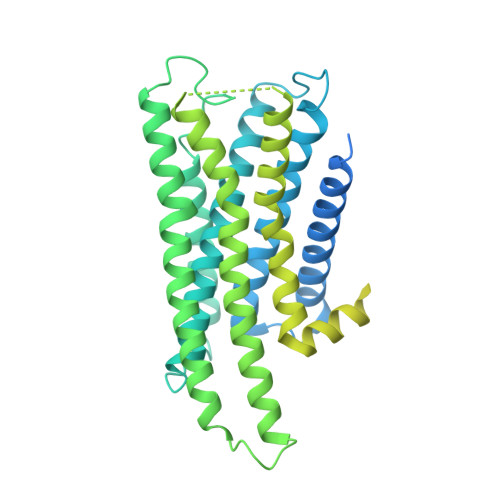
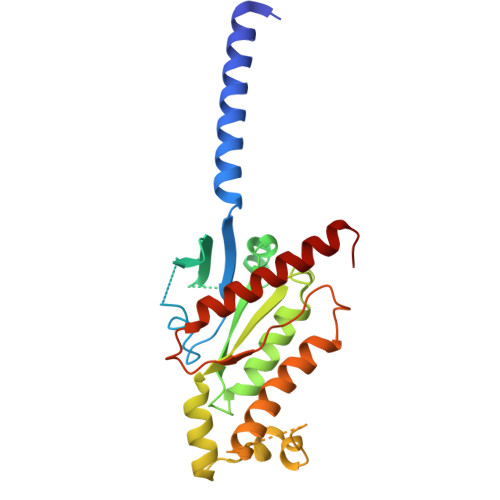
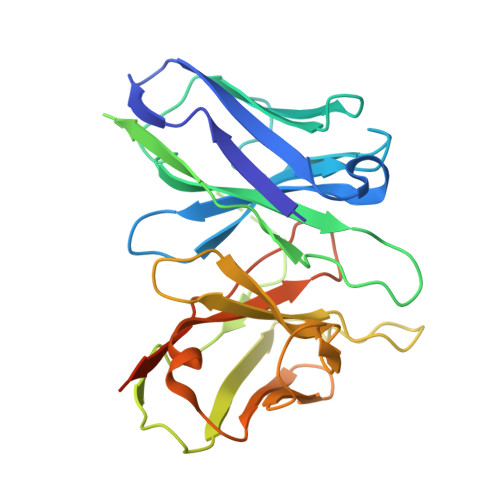
Divergent activation patterns of BRS3 revealed by two Chinese herb-derived agonists.
Li, J., Li, C., Zhou, Q., Han, W., Fang, M., Xu, Y., Mai, Y., Zhang, Y., Cui, J., Xu, H.E., Zhang, Y., Yin, W., Wang, M.W.(2025) Acta Pharm Sin B 15: 5231-5243
- PubMed: 41132856
- DOI: https://doi.org/10.1016/j.apsb.2025.06.025
- Primary Citation of Related Structures:
9K07, 9LWP - PubMed Abstract:
Bombesin receptor subtype-3 (BRS3) is an orphan G protein-coupled receptor (GPCR) that plays critical roles in energy homeostasis, glucose metabolism, and insulin secretion. Recent structural studies have elucidated BRS3 signaling mechanisms using synthetic ligands, including BA1 and MK-5046. However, the molecular basis of BRS3 activation by bioactive natural compounds and their derivatives, particularly those derived from traditional Chinese medicine, remains unclear. Here, we present high-resolution cryogenic electron microscopy (cryo-EM) structures of the human BRS3-G q complex in both unliganded and active states bound by two herb-derived compounds (DSO-5a and oridonin), at resolutions of 2.9, 2.8, and 2.9 Å, respectively. These structures display distinct ligand recognition patterns between DSO-5a and oridonin. Although both compounds bind to the orthosteric pocket, they differentially engage the interaction network of BRS3, as demonstrated by mutagenesis studies assessing calcium mobilization and inositol phosphate 1 (IP1) accumulation. These findings enhance our understanding of BRS3 activation and provide valuable insights into the development of small-molecule BRS3 modulators with therapeutic potential.
- Department of Pharmacology, School of Basic Medical Sciences, Fudan University, Shanghai 200032, China.
Organizational Affiliation: