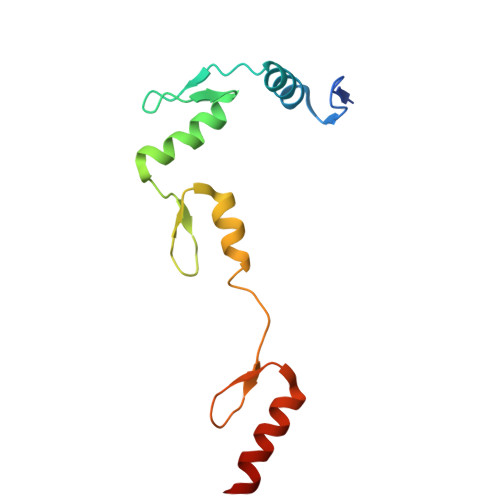
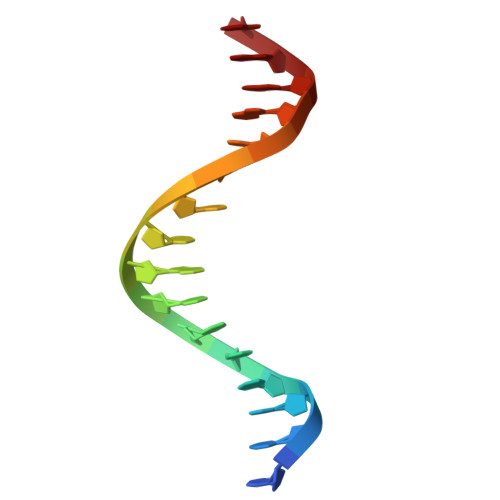
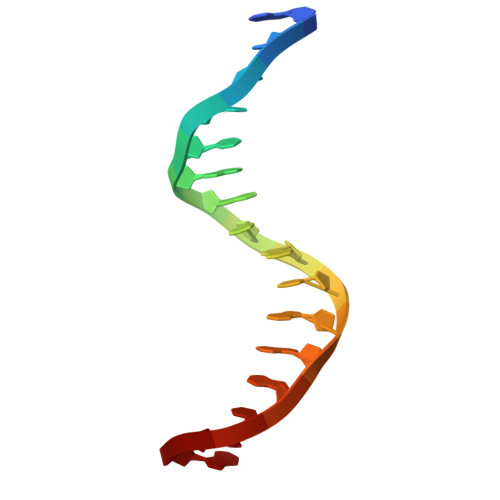
Structural insights into ZBTB20 action at the AFP promoter.
Yang, L., Xie, H., Wan, X., Li, M., Lv, M., Duan, Y., Shi, Y., Zhang, W.J., Li, F.(2025) Structure 33: 1398-1407.e2
- PubMed: 40494351
- DOI: https://doi.org/10.1016/j.str.2025.05.009
- Primary Citation of Related Structures:
9JZT - PubMed Abstract:
ZBTB20, a C2H2 zinc finger and broad-complex, tramtrack and bric-à-brac (BTB) domain-containing protein, is crucial for organ development and metabolic homeostasis. Its functionality is dependent on its DNA-binding zinc fingers, and heterozygous mutations within these regions are linked to Primrose syndrome, which is characterized by various physical and developmental abnormalities. However, the molecular basis underlying ZBTB20 zinc finger recognition of DNA remains largely unknown. Here, we present the crystal structure of ZBTB20 zinc fingers 1-4 (ZF1-4) in complex with the mouse alpha-fetoprotein (AFP) promoter in the region spanning positions -104 to -90. In combination with calorimetric analysis, we established that ZF1-3 is essential for the recognition of the AFP promoter and identified key residues involved in DNA binding. Furthermore, our data allow us to correlate Primrose syndrome mutations with alterations in DNA-binding efficacy. Overall, our study provides mechanistic insights into the physiological and pathological roles of ZBTB20 zinc fingers.
- MOE Key Laboratory for Cellular Dynamics, Hefei National Laboratory for Physical Sciences at the Microscale, The First Affiliated Hospital of USTC, Biomedical Sciences and Health Laboratory of Anhui Province, Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei 230027, China. Electronic address: ylna@ustc.edu.cn.
Organizational Affiliation: