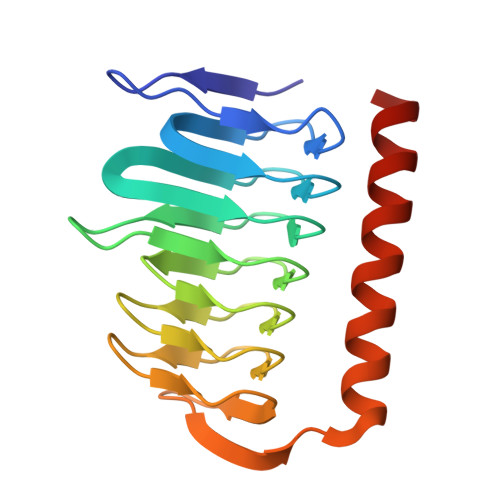
Crystal structure of gamma-carbonic anhydrase from the polyextremophilic bacterium Aeribacillus pallidus.
Choi, S.H., Jin, M.S.(2024) Mol Cells 48: 100165-100165
- PubMed: 39637945
- DOI: https://doi.org/10.1016/j.mocell.2024.100165
- Primary Citation of Related Structures:
9JYW - PubMed Abstract:
The polyextremophilic bacterium Aeribacillus pallidus produces a thermo- and alkali-stable γ-carbonic anhydrase (γ-apCA), a homotrimeric metalloenzyme containing a zinc ion in its active site that catalyzes the reversible hydration of carbon dioxide (CO 2 ). Here, we present the first crystal structure of γ-apCA at 1.7-Å resolution, revealing 2 trimers in the asymmetric unit. The overall structure is consistent with other γ-CAs, where each monomer adopts a prism-like structure consisting of an N-terminal left-handed β-helix and a C-terminal α-helix. The active site, located at the interface between 2 monomers, coordinates the zinc ion with 3 histidine residues (H65, H82, and H87) and a water molecule in a tetrahedral configuration. The structural comparison indicates that the amino acid composition at the active site of γ-apCA differs significantly from the prototypic γ-CA from Methanosarcina thermophila. This variation likely accounts for the lack of measurable CO 2 hydration activity in γ-apCA. Additionally, the structure reveals noncatalytic zinc and sulfate ions trapped at the trimer core and trimer-trimer noncrystallographic interfaces. These may contribute to stabilizing enzyme assembly and promoting crystal packing.
- School of Life Sciences, GIST, Gwangju 61005, Republic of Korea.
Organizational Affiliation: