Structures of the apo and agonist-crosslinked human gamma delta T-cell receptor
Wang, L., Li, Z., Fan, Y., Li, J.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| T-cell surface glycoprotein CD3 zeta chain | A [auth a], B [auth b] | 31 | Homo sapiens | Mutation(s): 0 Gene Names: CD247, CD3Z, T3Z, TCRZ | 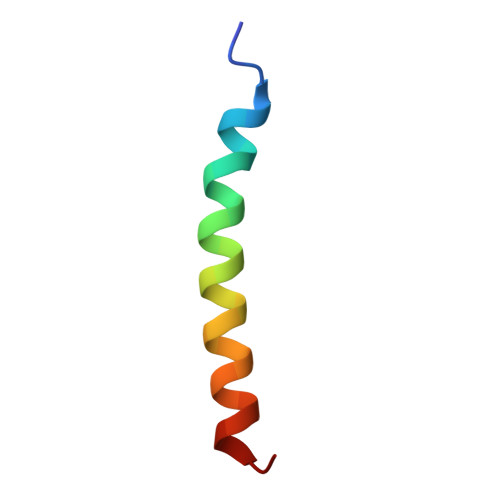 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P20963 (Homo sapiens) Explore P20963 Go to UniProtKB: P20963 | |||||
PHAROS: P20963 GTEx: ENSG00000198821 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P20963 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| T-cell surface glycoprotein CD3 delta chain | C [auth d] | 105 | Homo sapiens | Mutation(s): 0 Gene Names: CD3D, T3D | 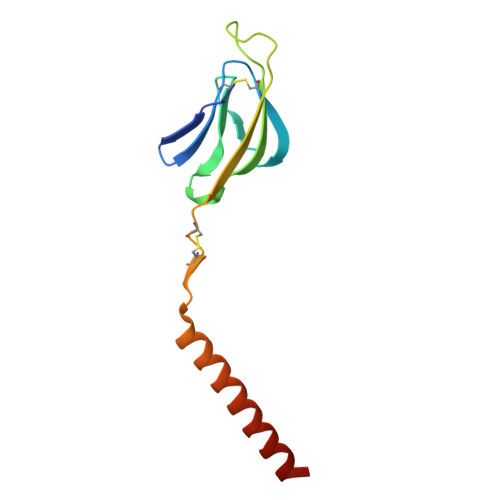 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P04234 (Homo sapiens) Explore P04234 Go to UniProtKB: P04234 | |||||
PHAROS: P04234 GTEx: ENSG00000167286 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P04234 | ||||
Glycosylation | |||||
| Glycosylation Sites: 2 | Go to GlyGen: P04234-1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| T-cell surface glycoprotein CD3 epsilon chain | D [auth e], E [auth f] | 122 | Homo sapiens | Mutation(s): 0 Gene Names: CD3E, T3E | 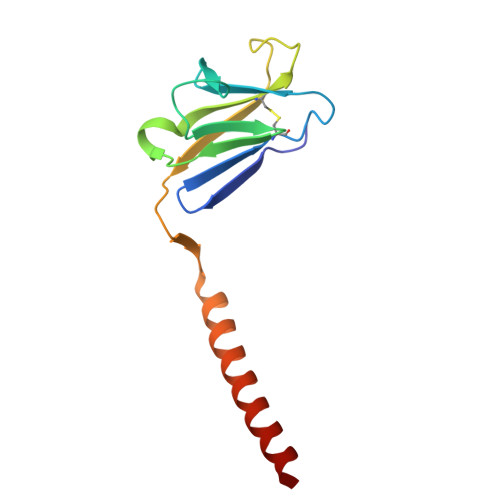 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P07766 (Homo sapiens) Explore P07766 Go to UniProtKB: P07766 | |||||
PHAROS: P07766 GTEx: ENSG00000198851 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P07766 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| T-cell surface glycoprotein CD3 gamma chain | F [auth g] | 115 | Homo sapiens | Mutation(s): 0 Gene Names: CD3G, T3G | 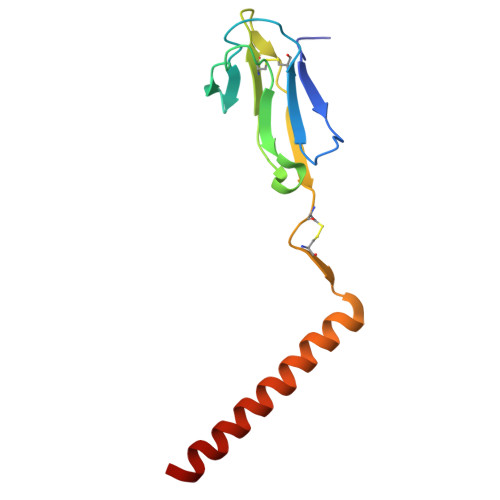 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P09693 (Homo sapiens) Explore P09693 Go to UniProtKB: P09693 | |||||
PHAROS: P09693 GTEx: ENSG00000160654 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P09693 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| T cell receptor delta constant | G [auth m] | 36 | Homo sapiens | Mutation(s): 0 Gene Names: TRDC |  |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for B7Z8K6 (Homo sapiens) Explore B7Z8K6 Go to UniProtKB: B7Z8K6 | |||||
PHAROS: B7Z8K6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | B7Z8K6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 6 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| T cell receptor gamma constant 1 | H [auth n] | 38 | Homo sapiens | Mutation(s): 0 Gene Names: TRGC1, TCRGC1 |  |
UniProt | |||||
Find proteins for P0CF51 (Homo sapiens) Explore P0CF51 Go to UniProtKB: P0CF51 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0CF51 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 7 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Fab light chain | I [auth A] | 104 | Homo sapiens | Mutation(s): 0 |  |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 8 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Fab heavy chain | J [auth B] | 121 | Homo sapiens | Mutation(s): 0 |  |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| CLR (Subject of Investigation/LOI) Query on CLR | K [auth b] | CHOLESTEROL C27 H46 O HVYWMOMLDIMFJA-DPAQBDIFSA-N |  | ||
| NAG (Subject of Investigation/LOI) Query on NAG | L [auth d], M [auth d] | 2-acetamido-2-deoxy-beta-D-glucopyranose C8 H15 N O6 OVRNDRQMDRJTHS-FMDGEEDCSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | |
| RECONSTRUCTION | cryoSPARC |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Not funded | -- |