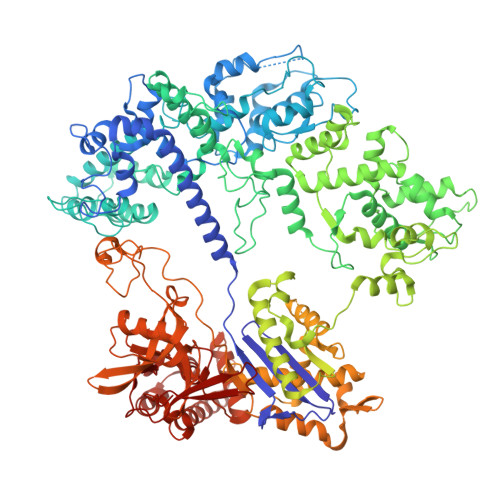
Structural and functional bases of F. rodentium Cas9 provide insights into CRISPR-Cas protein engineering.
Yang, M., Liu, S., Chen, G., Liu, X., Sun, D., Zhang, J., Wang, Y., Chen, S., Tian, R., Hu, Z.(2026) Cell Genom 6: 101039-101039
- PubMed: 41106392
- DOI: https://doi.org/10.1016/j.xgen.2025.101039
- Primary Citation of Related Structures:
9JWK, 9JWN - PubMed Abstract:
The Faecalibaculum rodentium (Fr) CRISPR-Cas9 system exhibits enhanced gene-editing precision and efficiency compared to SpCas9, with distinctive advantages in targeting the TATA box in eukaryotic promoters. However, the underlying molecular mechanisms remained unexplored. Here, we present cryo-electron microscopy structures of the FrCas9-single guide RNA (sgRNA)-DNA complex in both the R-loop expansion and pre-catalytic states, shedding light on its specialized recognition of the 5'-NRTA-3' protospacer adjacent motif (PAM) and the unusual overwinding of the sgRNA-DNA heteroduplex. Our investigations into the structure and extensive mutational analyses reveal that the phosphate lock loop plays a pivotal role in finely adjusting FrCas9's off-target sensitivity and catalytic efficiency. Remarkably, targeted residue substitutions in the phosphate lock loop and the PAM-distal region were found to synergistically enhance both the editing precision and efficiency of FrCas9. These findings advance our understanding of Cas9's accuracy and potency mechanisms while providing a molecular foundation for the rational design and development of next-generation CRISPR technologies.
- Department of Infectious Diseases, The Fifth Affiliated Hospital, Sun Yat-sen University, Zhuhai 519000, Guangdong, China; Molecular Imaging Center, The Fifth Affiliated Hospital, Sun Yat-sen University, Zhuhai 519000, Guangdong, China; Guangdong-Hong Kong-Macao University Joint Laboratory of Interventional Medicine, The Fifth Affiliated Hospital, Sun Yat-sen University, Zhuhai 519000, Guangdong, China.
Organizational Affiliation: