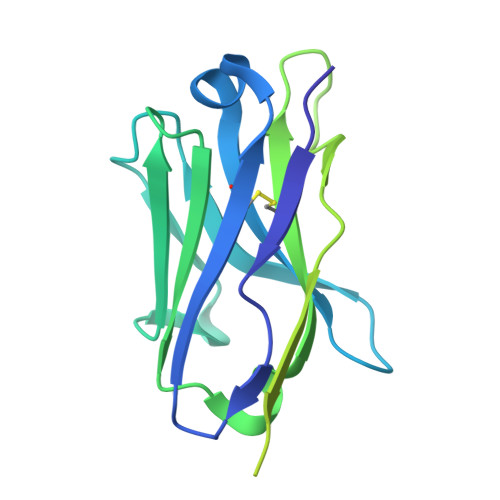
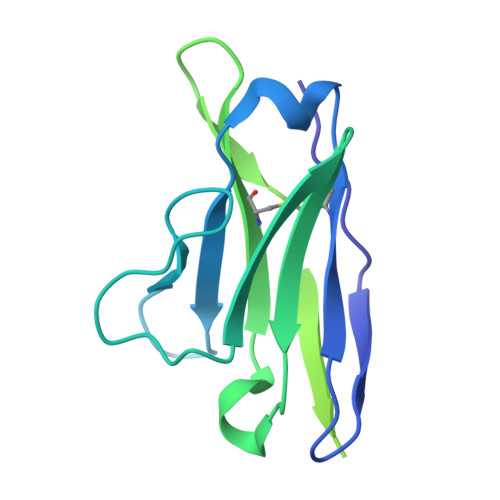
Structural polymorphism of the antigenic loop in HBV surface antigen dictates binding of diverse neutralizing antibodies.
He, X., Tao, W., Kang, Y., Xu, J., Liu, X., Chen, L.(2025) Cell Discov 11: 57-57
- PubMed: 40523894
- DOI: https://doi.org/10.1038/s41421-025-00803-2
- Primary Citation of Related Structures:
9IYY, 9JT1, 9U9B - PubMed Abstract:
The Hepatitis B Virus (HBV) poses a significant health threat, causing millions of deaths each year. Hepatitis B surface antigen (HBsAg), the sole membrane protein on the HBV viral envelope, plays crucial roles in viral attachment to host cells and serves as the target for neutralizing antibodies (NAbs). Despite its functional and therapeutic significance, the mechanisms by which NAbs recognize HBsAg remain elusive. Here, we found that HBsAg proteins exist in distinct subtypes and are recognized by different groups of antibodies. Cryo-electron microscopy (Cryo-EM) structures of HBsAg dimers in complex with NAb Fab fragments reveal that the antigenic loop (AGL) of these distinct HBsAg types share a common structural core comprised of four β-strands. However, their surface structures exhibit unexpected polymorphism due to distinct disulfide bond linkages within the AGL region. This structural polymorphism determines the recognition of HBsAg by different groups of NAbs.
- State Key Laboratory of Membrane Biology, College of Future Technology, Institute of Molecular Medicine, Beijing Key Laboratory of Cardiometabolic Molecular Medicine, Peking University, Beijing, China.
Organizational Affiliation: