Structure of endo-1.3-fucanase (Fun168E) at 2.04 Angstroms resulution.
Chen, G.N., Chang, Y.G.To be published.
Experimental Data Snapshot
Starting Model: in silico
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Glycoside-hydrolase family GH114 TIM-barrel domain-containing protein | 394 | Wenyingzhuangia fucanilytica | Mutation(s): 0 Gene Names: AXE80_08855 | 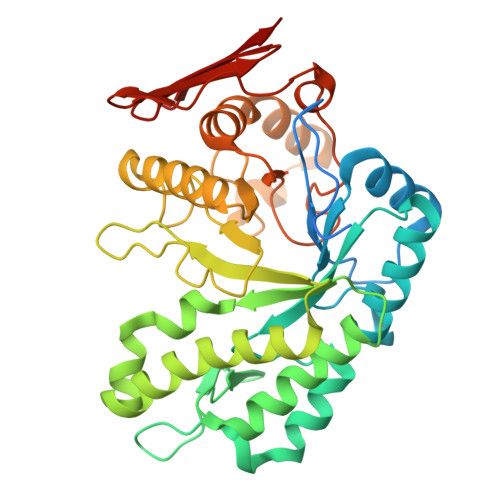 | |
UniProt | |||||
Find proteins for A0A1B1Y6K9 (Wenyingzhuangia fucanilytica) Explore A0A1B1Y6K9 Go to UniProtKB: A0A1B1Y6K9 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1B1Y6K9 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 83.135 | α = 90 |
| b = 83.135 | β = 90 |
| c = 100.43 | γ = 120 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| Aimless | data scaling |
| XDS | data reduction |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Natural Science Foundation of China (NSFC) | China | U22A20542 |