Molecular Mechanisms of Methamphetamine-Induced Addiction via TAAR1 Activation.
Lin, Y., Wang, J., Shi, F., Yang, L., Wu, S., Qiao, A., Ye, S.(2024) J Med Chem 67: 18593-18605
- PubMed: 39358311
- DOI: https://doi.org/10.1021/acs.jmedchem.4c01961
- Primary Citation of Related Structures:
9JKQ - PubMed Abstract:
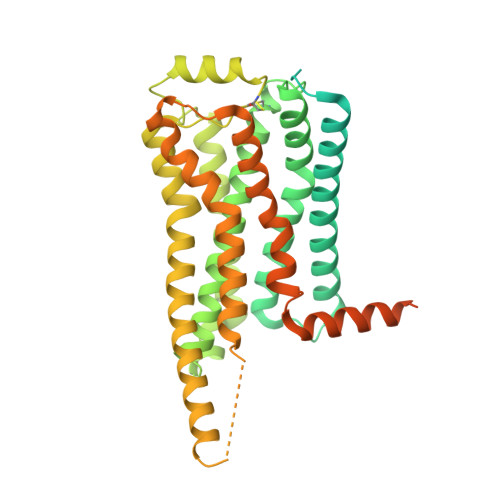
Trace amine-associated receptor 1 (TAAR1), a member of the trace amine receptor family, recognizes various trace amines in the brain, including endogenous β-phenylethylamine (PEA) and methamphetamine (METH). TAAR1 is a novel target for several neurological disorders, including schizophrenia, depression, and substance abuse. Herein, we report the structure of the human TAAR1-G s protein complex bound to METH. Using functional studies, we reveal the molecular basis of METH recognition by TAAR1, and potential mechanisms underlying the selectivity of TAAR1 for different ligands. Molecular dynamics simulations further elucidated possible mechanisms for the binding of chiral amphetamine (AMPH)-like psychoactive drugs to TAAR1. Additionally, we discovered a hydrophobic core on the transmembrane helices (TM), TM5 and TM6, explaining the unique mechanism of TAAR1 activation. These findings reveal the ligand recognition pattern and activation mechanism of TAAR1, which has important implications for the development of next-generation treatments for substance abuse and various neurological disorders.
- Tianjin Key Laboratory of Function and Application of Biological Macromolecular Structures, School of Life Sciences, Tianjin University, 92 Weijin Road, Nankai District, Tianjin 300072, China.
Organizational Affiliation: