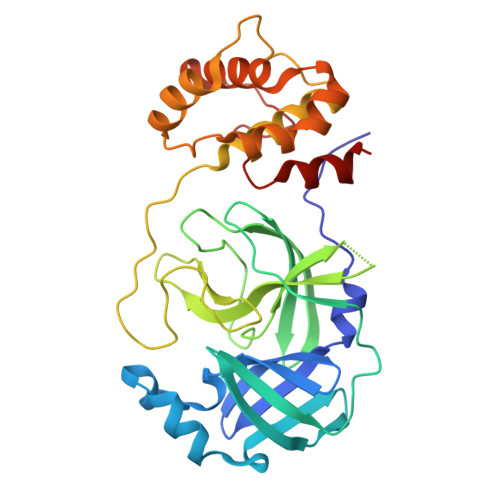
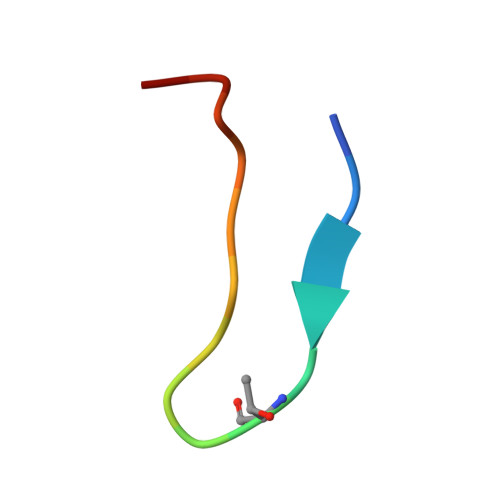
De novo discovery of a non-covalent cell-penetrating bicyclic pep-tide inhibitor targeting SARS-CoV-2 main protease
Yin, Y.Y., Shi, Y.S., Qi, J.Q., Tan, Y.T., Yang, J.Y., Wang, M.W., Peng, Q.P., Li, Y.L., Fu, L.F., Zhang, M.Z., Zhang, Y.Z.To be published.