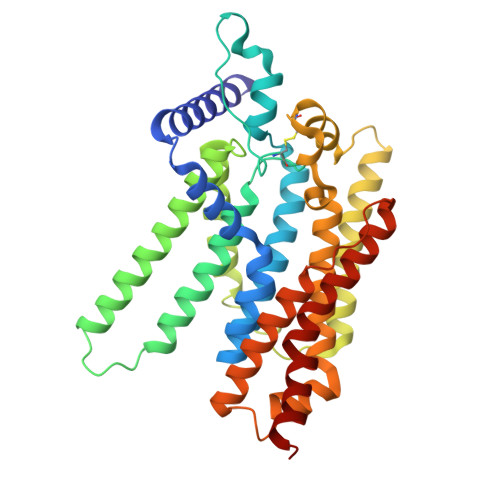
Structural insights into glucose-6-phosphate recognition and hydrolysis by human G6PC1.
Xia, Z., Liu, C., Wu, D., Chen, H., Zhao, J., Jiang, D.(2025) Proc Natl Acad Sci U S A 122: e2418316122-e2418316122
- PubMed: 39847333
- DOI: https://doi.org/10.1073/pnas.2418316122
- Primary Citation of Related Structures:
9J7U, 9J7V - PubMed Abstract:
The glucose-6-phosphatase (G6Pase) is an integral membrane protein that catalyzes the hydrolysis of glucose-6-phosphate (G6P) in the endoplasmic reticulum lumen and plays a vital role in glucose homeostasis. Dysregulation or genetic mutations of G6Pase are associated with diabetes and glycogen storage disease 1a (GSD-1a). Studies have characterized the biophysical and biochemical properties of G6Pase; however, the structure and substrate recognition mechanism of G6Pase remain unclear. Here, we present two cryo-EM structures of the 40-kDa human G6Pase: a wild-type apo form and a mutant G6Pase-H176A with G6P bound, elucidating the structural basis for substrate recognition and hydrolysis. G6Pase comprises nine transmembrane helices and possesses a large catalytic pocket facing the lumen. Unexpectedly, G6P binding induces substantial conformational rearrangements in the catalytic pocket, which facilitate the binding of the sugar moiety. In conjunction with functional analyses, this study provides critical insights into the structure, substrate recognition, catalytic mechanism, and pathology of G6Pase.
- Beijing National Laboratory for Condensed Matter Physics, Laboratory of Soft Matter Physics, Institute of Physics, Chinese Academy of Sciences, Beijing 100190, China.
Organizational Affiliation: