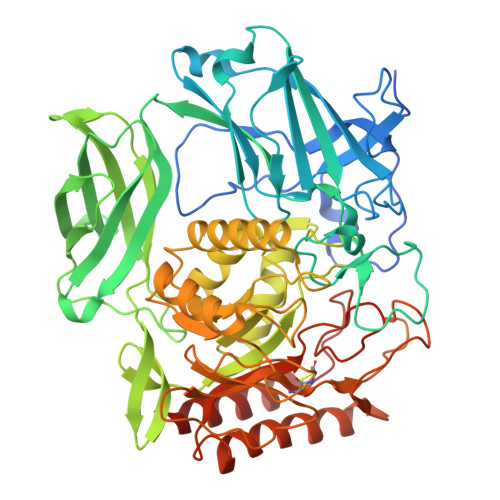
Crystal structure of beta-d-galactofuranosidase from Streptomyces sp. JHA19 in complex with an inhibitor provides insights into substrate specificity.
Fujio, N., Yamada, C., Kashima, T., Matsunaga, E., Nash, R.J., Takegawa, K., Fushinobu, S.(2024) FEBS Lett 598: 2866-2875
- PubMed: 39543437
- DOI: https://doi.org/10.1002/1873-3468.15056
- Primary Citation of Related Structures:
9J6M, 9J6N - PubMed Abstract:
d-Galactofuranose (Galf) is widely distributed in glycoconjugates of pathogenic microbes. β-d-Galactofuranosidase (Galf-ase) from Streptomyces sp. JHA19 (ORF1110) belongs to glycoside hydrolase (GH) family 2 and is the first identified Galf-specific degradation enzyme. Here, the crystal structure of ORF1110 in complex with a mechanism-based potent inhibitor, d-iminogalactitol (K i = 65 μm) was solved. ORF1110 binds to the C5-C6 hydroxy groups of d-iminogalactitol with an extensive and integral hydrogen bond network, a key interaction that discriminates the substrates. The active site structure of ORF1110 is largely different from those of β-glucuronidases and β-galactosidases in the same GH2 family. A C-terminal domain of ORF1110 is predicted to be a carbohydrate-binding module family 42 that may bind Galf. The structural insights into Galf-ase will contribute to the investigation of therapeutic tools against pathogens.
- Department of Biotechnology, The University of Tokyo, Japan.
Organizational Affiliation: