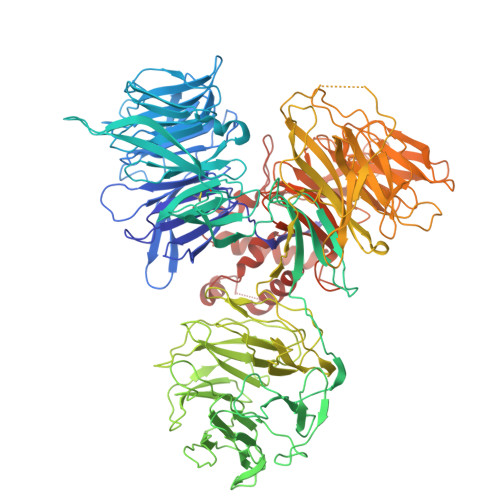
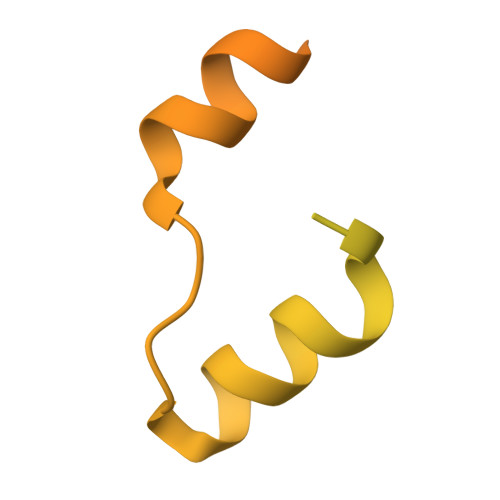
Structural basis of the hepatitis B virus X protein in complex with DDB1.
Tanaka, H., Diogo Dias, J., Jay, B., Kita, S., Sasaki, M., Takeda, H., Kishimoto, N., Sasaki, S., Misumi, S., Mizokami, M., Neuveut, C., Sumikama, T., Shibata, M., Maenaka, K., Machida, S.(2025) Proc Natl Acad Sci U S A 122: e2421325122-e2421325122
- PubMed: 40512786
- DOI: https://doi.org/10.1073/pnas.2421325122
- Primary Citation of Related Structures:
9J6J, 9J6K - PubMed Abstract:
A cure for chronic hepatitis B requires eliminating or permanently silencing covalently closed circular DNA (cccDNA). A pivotal target of this approach is the hepatitis B virus (HBV) X protein (HBx), which is a key factor that promotes transcription from cccDNA. However, the HBx structure remains unsolved. Here, we present the cryoelectron microscopy structure of HBx in complex with DDB1, which is an essential complex for cccDNA transcription. In this structure, hydrophobic interactions within HBx were identified, and mutational analysis highlighted their importance in the HBV life cycle. Our biochemical analysis revealed that the HBx-DDB1 complex directly interacts simultaneously with NSE3, which is a component of the SMC5/6 complex, and Spindlin1. Additionally, HBx-DDB1 complex dynamics were explored via high-speed atomic force microscopy. These findings provide comprehensive insights into the structure and function of HBx in HBV replication.
- Department of Structural Virology, National Institute of Global Health and Medicine, Japan Institute for Health Security, Shinjuku-ku, Tokyo 162-8655, Japan.
Organizational Affiliation: