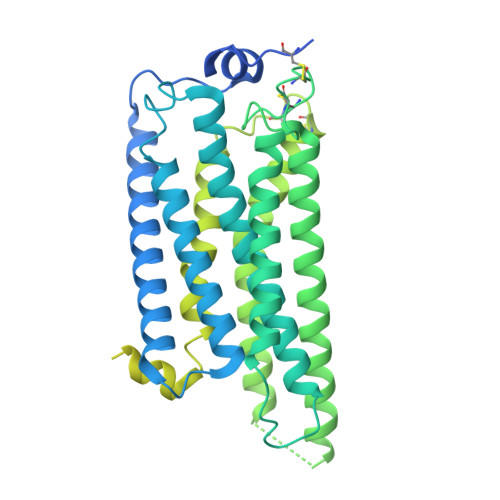
Structural mechanisms of potent lysophosphatidic acid receptor 1 activation by nonlipid basic agonists.
Akasaka, H., Sano, F.K., Shihoya, W., Nureki, O.(2024) Commun Biol 7: 1444-1444
- PubMed: 39506093
- DOI: https://doi.org/10.1038/s42003-024-07152-y
- Primary Citation of Related Structures:
9J5V - PubMed Abstract:
Lysophosphatidic acid receptor 1 (LPA 1 ) is one of the G protein-coupled receptors activated by the lipid mediator, lysophosphatidic acid (LPA). LPA 1 is associated with a variety of diseases, and LPA 1 agonists have potential therapeutic value for treating obesity and depression. Although potent nonlipid LPA 1 agonists have recently been identified, the mechanisms of nonlipid molecule-mediated LPA 1 activation remain unclear. Here, we report a cryo-electron microscopy structure of the human LPA 1 -G i complex bound to a nonlipid basic agonist, CpY, which has 30-fold higher agonistic activity as compared with LPA. Structural comparisons of LPA 1 with other lipid GPCRs revealed that the negative charge in the characteristic binding pocket of LPA 1 allows the selective recognition of CpY, which lacks a polar head. In addition, our structure show that the ethyl group of CpY directly pushes W271 6.48 to fix the active conformation. Endogenous LPA lacks these chemical features, which thus represent the crucial elements of nonlipid agonists that potently activate LPA 1 . This study provides detailed mechanistic insights into the ligand recognition and activation of LPA 1 by nonlipid agonists, expanding the scope for drug development targeting the LPA receptors.
- Department of Biological Sciences, Graduate School of Science, The University of Tokyo, Bunkyo, Tokyo, 113-0033, Japan.
Organizational Affiliation: