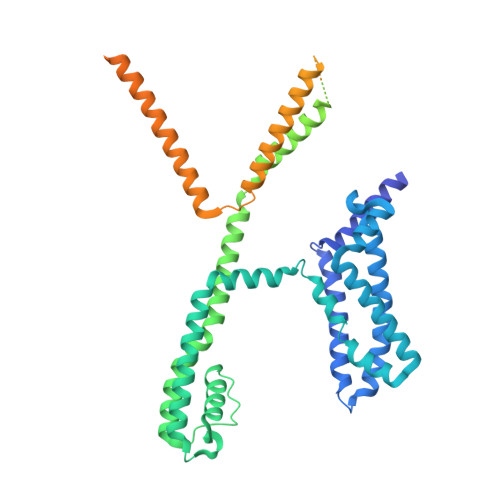
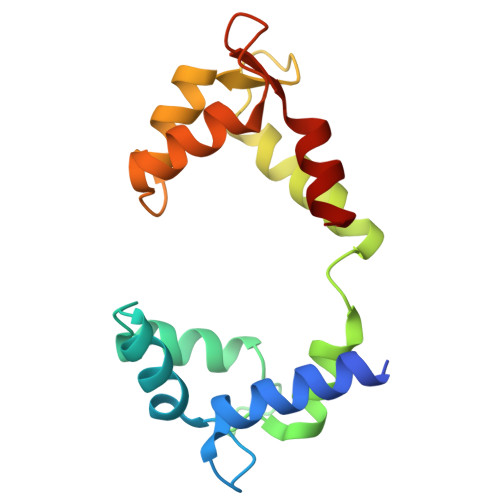
Phosphatidylinositol 4,5-bisphosphate activation mechanism of human KCNQ5.
Yang, Z., Zheng, Y., Ma, D., Wang, L., Zhang, J., Song, T., Wang, Y., Zhang, Y., Nan, F., Su, N., Gao, Z., Guo, J.(2025) Proc Natl Acad Sci U S A 122: e2416738122-e2416738122
- PubMed: 40172963
- DOI: https://doi.org/10.1073/pnas.2416738122
- Primary Citation of Related Structures:
9J38, 9LIZ, 9LJ1, 9LJ5 - PubMed Abstract:
The human voltage-gated potassium channels KCNQ2, KCNQ3, and KCNQ5 can form homo- and heterotetrameric channels that are responsible for generating the neuronal M current and maintaining the membrane potential stable. Activation of KCNQ channels requires both the depolarization of membrane potential and phosphatidylinositol 4,5-bisphosphate (PIP 2 ). Here, we report cryoelectron microscopy structures of the human KCNQ5-calmodulin (CaM) complex in the apo, PIP 2 -bound, and both PIP 2 - and the activator HN37-bound states in either a closed or an open conformation. In the closed conformation, a PIP 2 molecule binds in the middle of the groove between two adjacent voltage-sensing domains (VSDs), whereas in the open conformation, one additional PIP 2 binds to the interface of VSD and the pore domain, accompanying structural rearrangement of the cytosolic domain of KCNQ and CaM. The structures, along with electrophysiology analyses, reveal the two different binding modes of PIP 2 and elucidate the PIP 2 activation mechanism of KCNQ5.
- Department of Biophysics and Department of Neurology of the Fourth Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, Zhejiang 310058, China.
Organizational Affiliation: