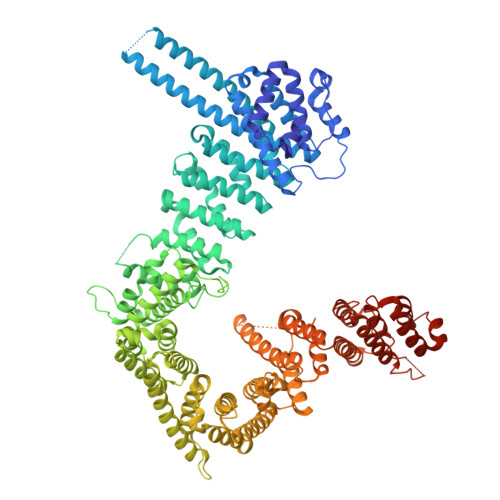
ANKRD11 binding to cohesin suggests a connection between KBG syndrome and Cornelia de Lange syndrome.
Liu, H., Li, H., Cai, Q., Zhang, J., Zhong, H., Hu, G., Zhao, S., Lu, Y., Mao, Y., Lu, Y., Yao, H., Zhang, M.(2025) Proc Natl Acad Sci U S A 122: e2417346122-e2417346122
- PubMed: 39847329
- DOI: https://doi.org/10.1073/pnas.2417346122
- Primary Citation of Related Structures:
9J0A - PubMed Abstract:
Ankyrin Repeat Domain-containing Protein 11 ( ANKRD11 ) is a causative gene for KBG syndrome, a significant risk factor for Cornelia de Lange syndrome (CdLS), and a highly confident autism spectrum disorder gene. Mutations of ANKRD11 lead to developmental abnormalities in multiple organs/tissues including the brain, craniofacial and skeletal bones, and tooth structures with unknown mechanism(s). Here, we find that ANKRD11, via a short peptide fragment in its N-terminal region, binds to the cohesin complex with a high affinity, implicating why ANKRD11 mutation can cause CdLS. The crystal structure of the ANKRD11 peptide in complex with cohesin, together with biochemical experiments, revealed that ANKRD11 competes with CCCTC-binding factor in binding to the cohesin complex. Importantly, a single point mutation in ANKRD11 (Tyr347 to Ala) specifically disrupted the interaction between ANKRD11 and cohesin and perturbed gene expressions in a mouse embryonic stem cell model. Mice carrying the ANKRD11 Y347A mutation display neural and craniofacial anomalies, which mirror clinical phenotypes observed in KBG syndrome patients. Thus, our study reveals how ANKRD11 functions together with cohesin to regulate gene expression and also provides insights into the molecular mechanisms underpinning developmental disorders caused by ANKRD11 mutations.
- Shenzhen Key Laboratory of Biomolecular Assembling and Regulation, Department of Neuroscience, School of Life Sciences, Southern University of Science and Technology, Shenzhen 518055, China.
Organizational Affiliation: