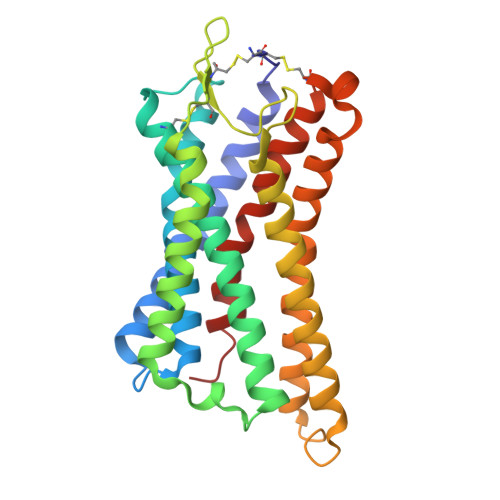
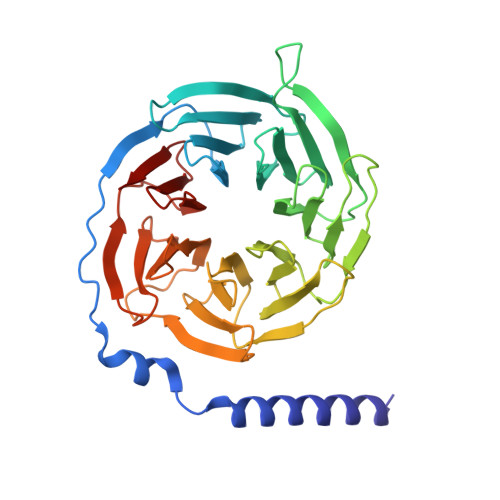
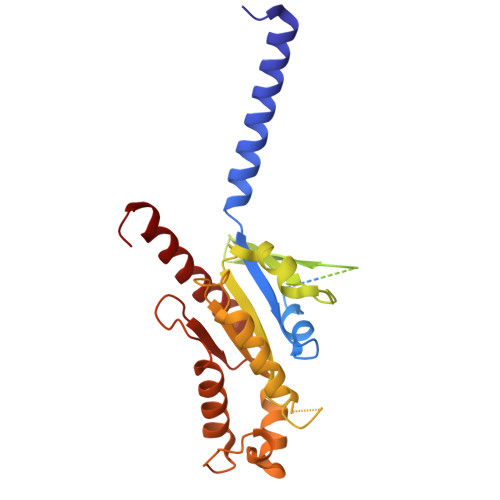
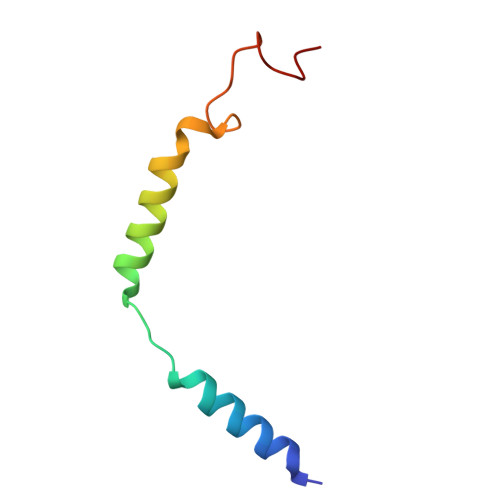
Structures of G-protein coupled receptor HCAR1 in complex with Gi1 protein reveal the mechanistic basis for ligand recognition and agonist selectivity.
Pan, X., Ye, F., Ning, P., Yu, Y., Zhang, Z., Wang, J., Chen, G., Wu, Z., Qiu, C., Li, J., Chen, B., Zhu, L., Qian, C., Gong, K., Du, Y.(2025) PLoS Biol 23: e3003126-e3003126
- PubMed: 40233099
- DOI: https://doi.org/10.1371/journal.pbio.3003126
- Primary Citation of Related Structures:
9IZA, 9IZC, 9IZD, 9J8Z - PubMed Abstract:
Hydroxycarboxylic acid receptor 1 (HCAR1), also known as lactate receptor or GPR81, is a class A G-protein-coupled receptor with key roles in regulating lipid metabolism, neuroprotection, angiogenesis, cardiovascular function, and inflammatory response in humans. HCAR1 is highly expressed in numerous types of cancer cells, where it participates in controlling cancer cell metabolism and defense mechanisms, rendering it an appealing target for cancer therapy. However, the molecular basis of HCAR1-mediated signaling remains poorly understood. Here, we report four cryo-EM structures of human HCAR1 and HCAR2 in complex with the Gi1 protein, in which HCAR1 binds to the subtype-specific agonist CHBA (3.16 Å) and apo form (3.36 Å), and HCAR2 binds to the subtype-specific agonists MK-1903 (2.68 Å) and SCH900271 (3.06 Å). Combined with mutagenesis and cellular functional assays, we elucidate the mechanisms underlying ligand recognition, receptor activation, and G protein coupling of HCAR1. More importantly, the key residues that determine ligand selectivity between HCAR1 and HCAR2 are clarified. On this basis, we further summarize the structural features of agonists that match the orthosteric pockets of HCAR1 and HCAR2. These structural insights are anticipated to greatly accelerate the development of novel HCAR1-targeted drugs, offering a promising avenue for the treatment of various diseases.
- Department of Cardiology, Institute of Cardiovascular Disease, Yangzhou Key Lab of Innovation Frontiers in Cardiovascular Disease, Affiliated Hospital of Yangzhou University, Yangzhou University, Yangzhou, China.
Organizational Affiliation: