cryo-EM structure of FtsE/X and ZipA complex in filament
Zhu, K.F., Li, J.W., Luo, M.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cell division protein ZipA | 328 | Escherichia coli str. K-12 substr. MG1655 | Mutation(s): 0 Gene Names: zipA, b2412, JW2404 | 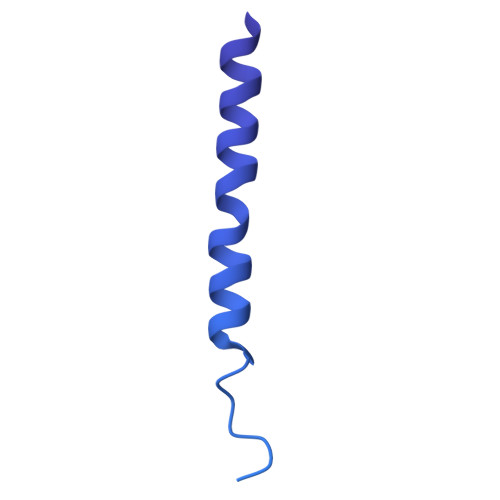 | |
UniProt | |||||
Find proteins for P77173 (Escherichia coli (strain K12)) Explore P77173 Go to UniProtKB: P77173 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P77173 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cell division ATP-binding protein FtsE | 226 | Escherichia coli str. K-12 substr. MG1655 | Mutation(s): 0 Gene Names: ftsE | 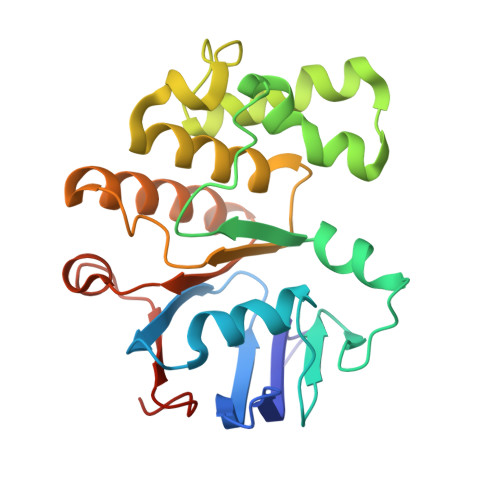 | |
UniProt | |||||
Find proteins for P0A9R7 (Escherichia coli (strain K12)) Explore P0A9R7 Go to UniProtKB: P0A9R7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0A9R7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cell division protein FtsX | 352 | Escherichia coli str. K-12 substr. MG1655 | Mutation(s): 0 Gene Names: ftsX, ftsS, b3462, JW3427 | 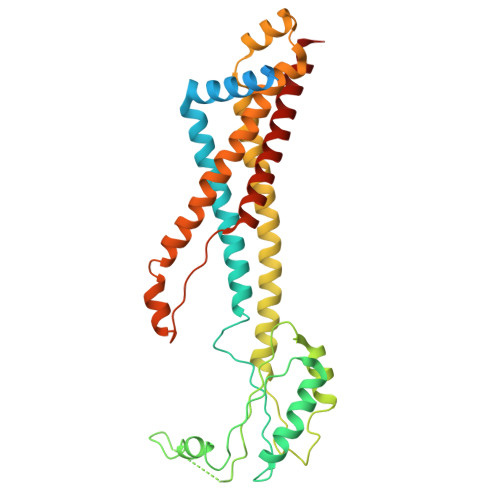 | |
UniProt | |||||
Find proteins for P0AC30 (Escherichia coli (strain K12)) Explore P0AC30 Go to UniProtKB: P0AC30 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0AC30 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| AGS (Subject of Investigation/LOI) Query on AGS | CB [auth 5] DB [auth 6] EB [auth G] FB [auth H] GB [auth M] | PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER C10 H16 N5 O12 P3 S NLTUCYMLOPLUHL-KQYNXXCUSA-N |  | ||
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Research Foundation (NRF, Singapore) | Singapore | -- |