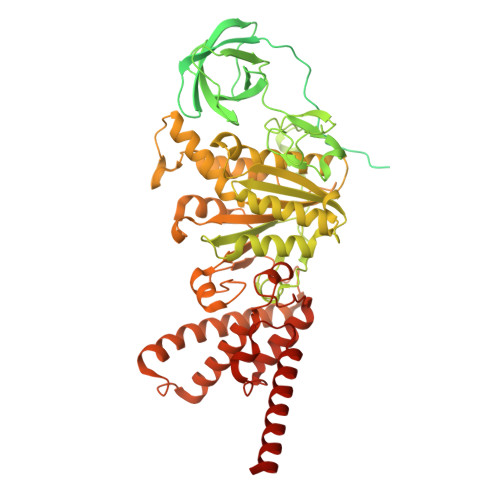
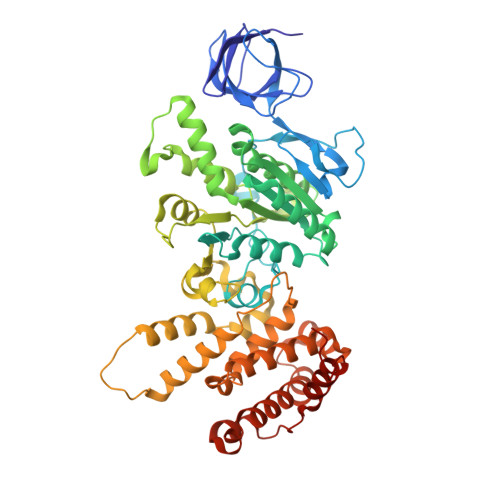
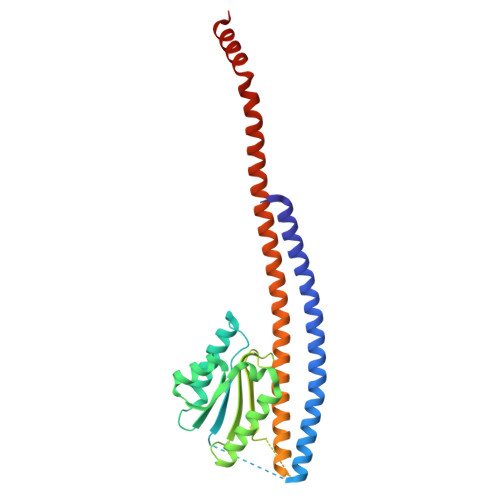
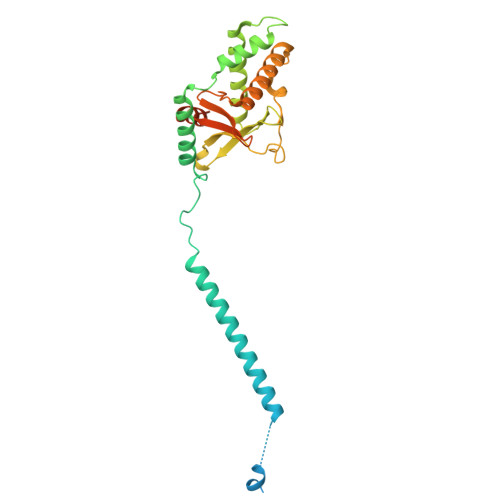
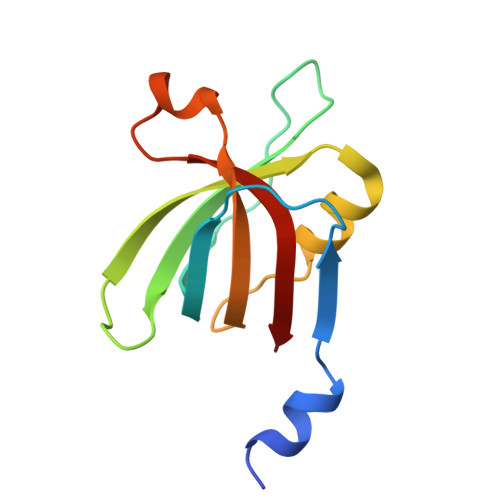
Dimeric assembly of F 1 -like ATPase for the gliding motility of Mycoplasma.
Toyonaga, T., Kato, T., Kawamoto, A., Miyata, T., Kawakami, K., Fujita, J., Hamaguchi, T., Namba, K., Miyata, M.(2025) Sci Adv 11: eadr9319-eadr9319
- PubMed: 40009674
- DOI: https://doi.org/10.1126/sciadv.adr9319
- Primary Citation of Related Structures:
9IO5 - PubMed Abstract:
Rotary ATPases, including F 1 F O -, V 1 V O -, and A 1 A O -ATPases, are molecular motors that exhibit rotational movements for energy conversion. In the gliding bacterium, Mycoplasma mobile , a dimeric F 1 -like ATPase forms a chain structure within the cell, which is proposed to drive the gliding motility. However, the mechanisms of force generation and transmission remain unclear. We determined the electron cryomicroscopy (cryo-EM) structure of the dimeric F 1 -like ATPase complex. The structure revealed an assembly distinct from those of dimeric F 1 F O -ATPases. The F 1 -like ATPase unit associated by two subunits GliD and GliE was named G 1 -ATPase as an R 1 domain of rotary ATPases. G 1 -β subunit, a homolog of the F 1 -ATPase catalytic subunit, exhibited a specific N-terminal region that incorporates the glycolytic enzyme, phosphoglycerate kinase into the complex. Structural features of the ATPase displayed strong similarities to F 1 -ATPase, suggesting a rotation based on the rotary catalytic mechanism. Overall, the cryo-EM structure provides insights into the mechanism through which G 1 -ATPase drives the Mycoplasma gliding motility.
- Graduate School of Science, Osaka Metropolitan University, 3-3-138 Sugimoto, Sumiyoshi-ku, Osaka 558-8585, Japan.
Organizational Affiliation: