A mutual co-recognition mechanism ensures the proper assembly of heterotrimeric kinesin-2 for intraflagellar transport.
Ren, J., Zhao, L., Chen, G., Ou, G., Feng, W.(2025) Nat Commun 16: 6816-6816
- PubMed: 40707480
- DOI: https://doi.org/10.1038/s41467-025-62152-8
- Primary Citation of Related Structures:
9IKB - PubMed Abstract:
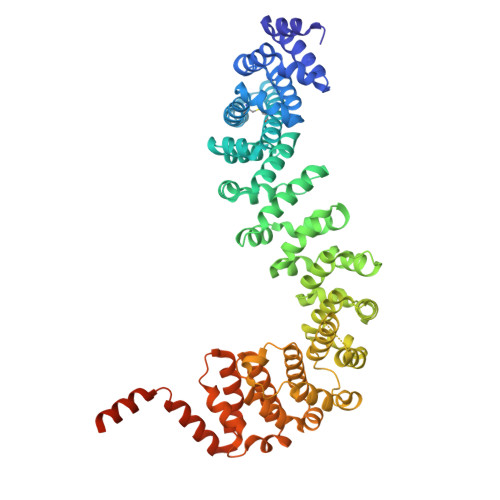
Heterotrimeric kinesin-2, composed of two distinct kinesin motors and a kinesin-associated protein (KAP), is essential for intraflagellar transport and ciliogenesis. KAP specifically recognizes the hetero-paired motor tails for the holoenzyme assembly, but the underlying mechanism remains unclear. Here, we determine the structure of KAP-1 in complex with the hetero-paired tails from kinesin-2 motors KLP-20 and KLP-11. KAP-1 forms an elongated superhelical structure characterized by a central groove and a C-terminal helical (CTH)-hook. The two motor tails fold together and are co-recognized by the central groove of KAP-1. The adjacent hetero-pairing trigger sequences preceding the two tails form an intertwined heterodimer, which co-captures the CTH-hook of KAP-1 to complete the holoenzyme assembly. Mutations in the interfaces between KAP-1 and the two tails disrupt the heterotrimeric kinesin-2 complex and impair kinesin-2-mediated intraflagellar transport. Thus, KAP-1 and the hetero-paired motors are mutually co-recognized, ensuring the proper assembly of heterotrimeric kinesin-2 for cargo transport.
- State Key Laboratory of Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing, China.
Organizational Affiliation: