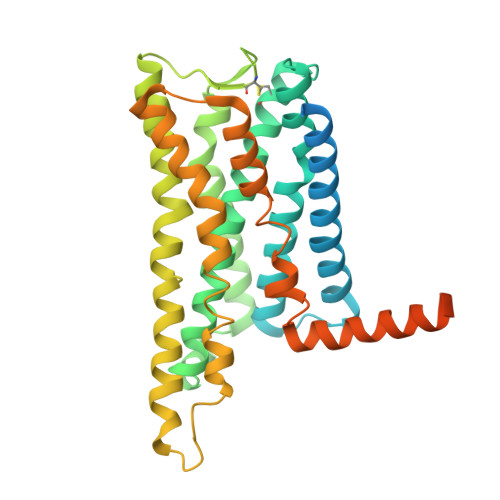
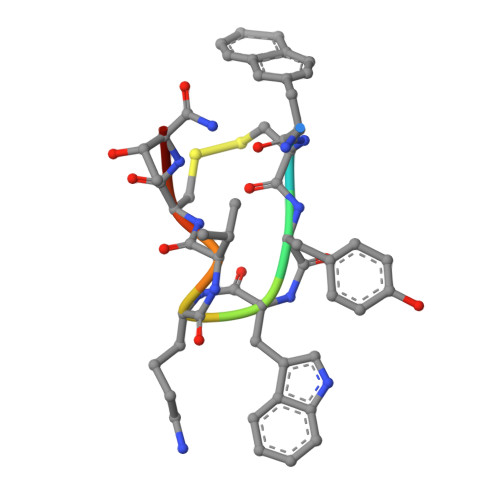
Structural insights into the binding modes of lanreotide and pasireotide with somatostatin receptor 1.
Zeng, Z., Liao, Q., Gan, S., Li, X., Xiong, T., Xu, L., Li, D., Jiang, Y., Chen, J., Ye, R., Du, Y., Wong, T.(2025) Acta Pharm Sin B 15: 2468-2479
- PubMed: 40487642
- DOI: https://doi.org/10.1016/j.apsb.2025.03.043
- Primary Citation of Related Structures:
9IK8, 9IK9 - PubMed Abstract:
Somatostatin receptor 1 (SSTR1) is a crucial therapeutic target for various neuroendocrine and oncological disorders. Current SSTR1-targeted treatments, including the first-generation somatostatin analog lanreotide (Lan) and the second-generation analog pasireotide (Pas), show promise but encounter challenges related to selectivity and efficacy. This study presents high-resolution cryo-electron microscopy structures of SSTR1 complexed with Lan or Pas, revealing the distinct mechanisms of ligand-binding and activation. These structures illustrate unique conformational changes in the SSTR1 orthosteric pocket induced by each ligand, which are critical for receptor activation and ligand selectivity. Combined with the biochemical assays and molecular dynamics simulations, our results provide a comparative analysis of binding characteristics within the SSTR family, highlighting subtle differences in SSTR1 activation by Lan and Pas. These insights pave the way for designing next-generation therapies with enhanced efficacy and reduced side effects through improved receptor subtype selectivity.
- Kobilka Institute of Innovative Drug Discovery, School of Medicine, the Chinese University of Hong Kong, Shenzhen 518172, China.
Organizational Affiliation: