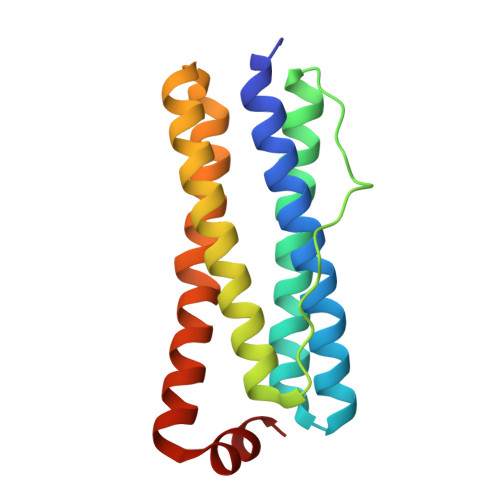
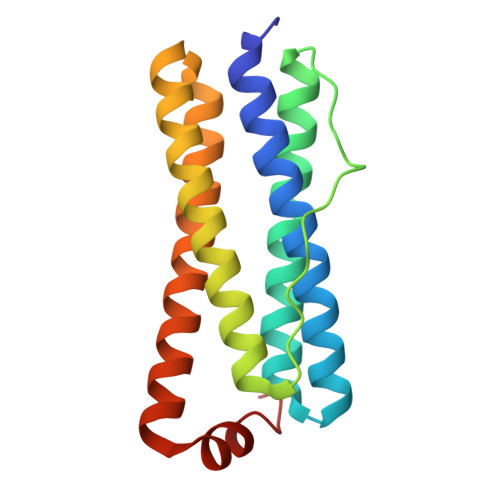
Unveiling Structural Heterogeneity and Evolutionary Adaptations of Heteromultimeric Bacterioferritin Nanocages.
Li, Y., Wang, W., Wang, W., Zhang, X., Chen, J., Gao, H.(2025) Adv Sci (Weinh) 12: e2409957-e2409957
- PubMed: 40167232
- DOI: https://doi.org/10.1002/advs.202409957
- Primary Citation of Related Structures:
9IIG - PubMed Abstract:
Iron-storage bacterioferritins (Bfrs), existing in either homo- or hetero-multimeric form, play a crucial role in iron homeostasis. While the structure and function of homo-multimeric bacterioferritins (homo-Bfrs) have been extensively studied, little is known about the assembly, distinctive characteristics, or evolutionary adaptations of hetero-multimeric bacterioferritins (hetero-Bfrs). Here, the cryo-EM structure and functional characterization of a bacterial hetero-Bfr (SoBfr12) are reported. Compared to homo-Bfrs, although SoBfr12 exhibits a conserved spherical cage-like dodecahedron, its pores through which ions traverse exhibit substantially increased diversity. Importantly, the heterogeneity has significant impacts on sites for ion entry, iron oxidation, and reduction. Moreover, evolutionary analyses reveal that hetero-Bfrs may represent a new class within the Bfr subfamily, consisting of two different types that may have evolved from homo-Bfr through tandem duplication and directly from ferritin (Ftn) via dispersed duplication, respectively. These results reveal remarkable structural and functional features of a hetero-Bfr, enabling the rational design of nanocages for enhanced iron-storing efficiency and for other specific purposes, such as drug delivery vehicles and nanozymes.
- State Key Laboratory for Vegetation Structure, Function and Construction (VegLab), Institute of Microbiology and College of Life Sciences, Zhejiang University, Hangzhou, 310058, China.
Organizational Affiliation: