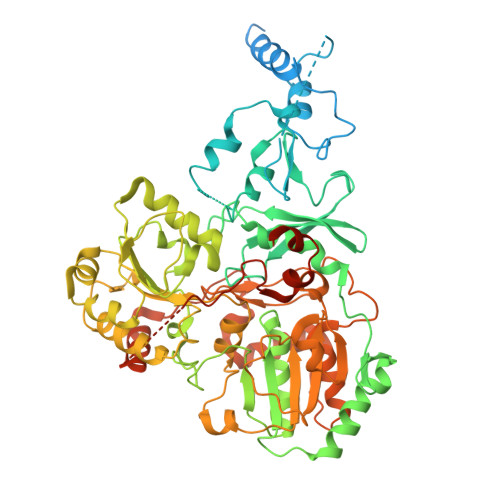
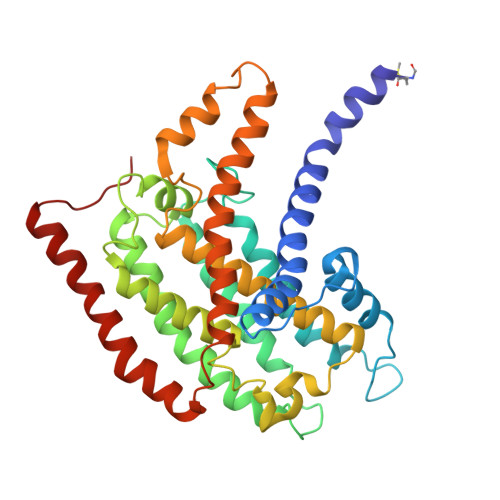
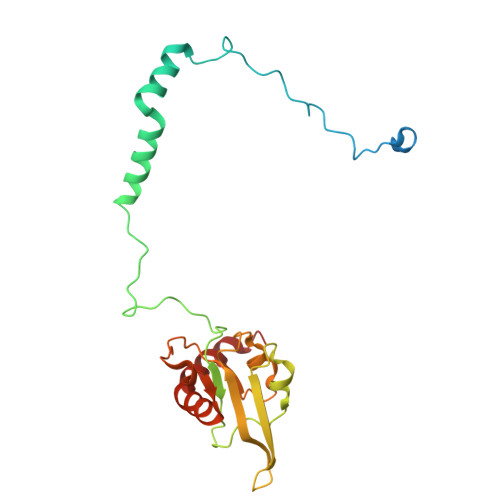
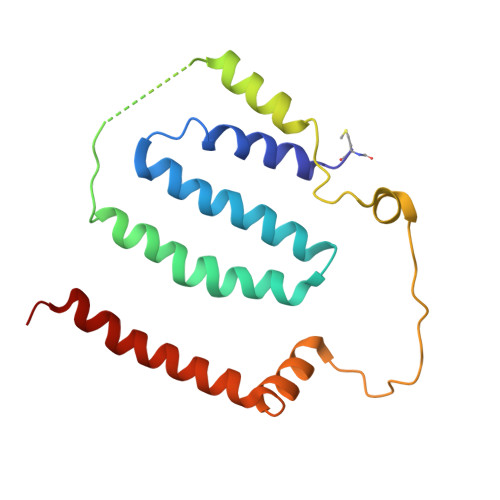
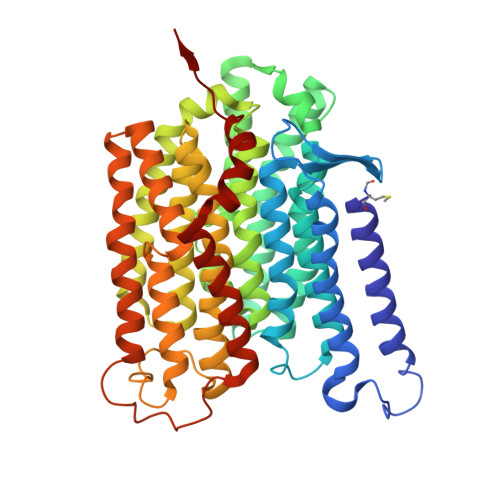
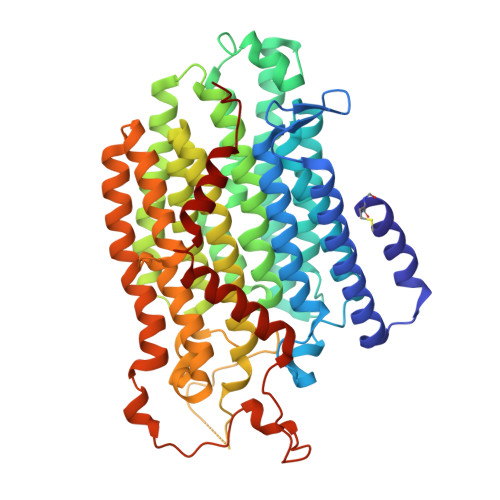
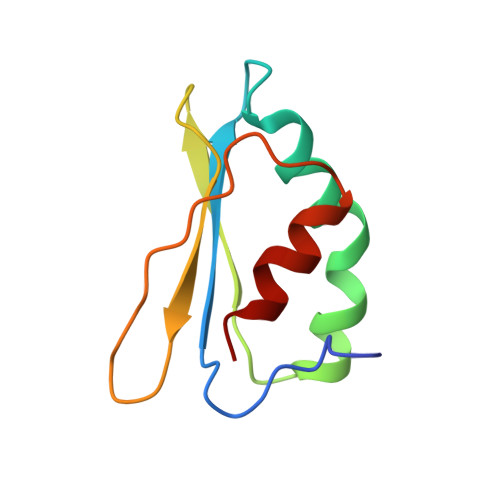
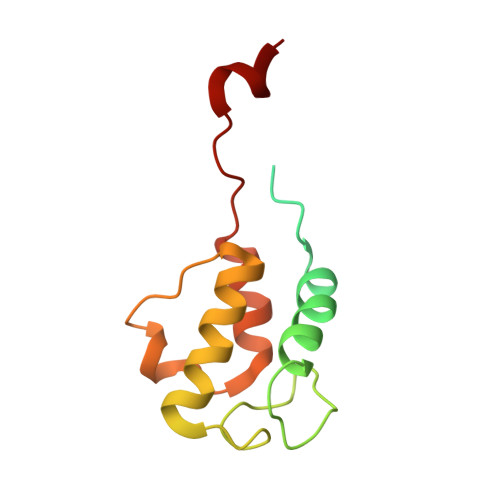
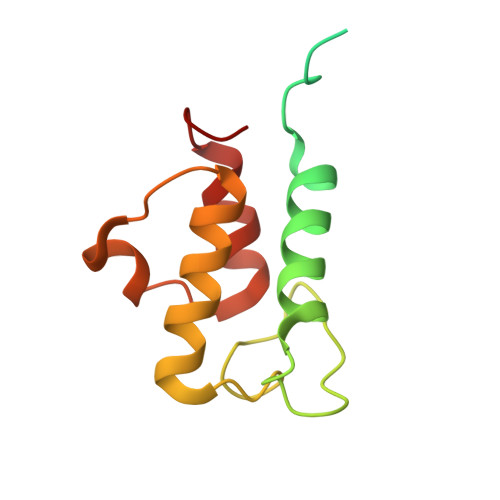
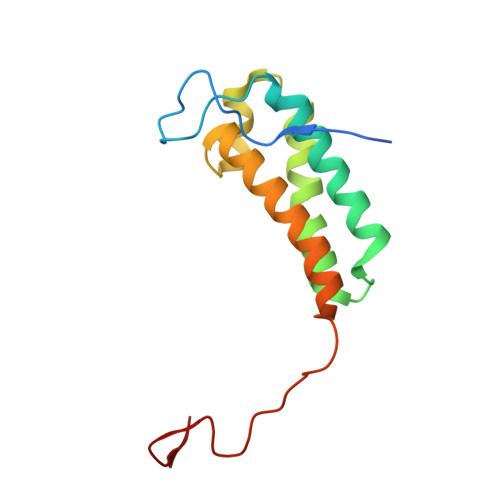
Global conformations of Pichia pastoris complex I are distinguished by the binding of a unique interdomain bridging subunit.
Lee, C.S., Grba, D.N., Wright, J.J., Ivanov, B.S., Hirst, J.(2025) Sci Adv 11: eadz0693-eadz0693
- PubMed: 41032597
- DOI: https://doi.org/10.1126/sciadv.adz0693
- Primary Citation of Related Structures:
9IHO, 9IHP, 9IHQ, 9IHR - PubMed Abstract:
Complex I (CI; NADH ubiquinone oxidoreductase) is central to energy generation and metabolic homeostasis in mammalian cells but contributes to adverse outcome pathways under challenging conditions. During ischemia, mammalian CI transitions from a turnover-ready, structurally "closed" state toward a dormant "open" state that prevents it from functioning in reverse during reperfusion to produce reactive oxygen species. Unfortunately, simpler, genetically tractable CI models do not recapitulate the same regulatory behavior, compromising mechanistic studies. Here, we report the structure of isolated CI from the yeast Pichia pastoris ( Pp -CI) and identify distinct closed and open states that resemble those of mammalian CI. Notably, a hitherto-unknown protein (NUQM) completes an interdomain bridge in only the closed state, implying that NUQM stabilizes it by restricting the conformational changes of opening. The direct correlation of NUQM binding with closed/open status in Pp -CI provides opportunities for investigating regulatory mechanisms relevant to reversible catalysis and ischemia-reperfusion injury.
- The Medical Research Council Mitochondrial Biology Unit, University of Cambridge, Keith Peters Building, Cambridge Biomedical Campus, Hills Road, Cambridge CB2 0XY, UK.
Organizational Affiliation: