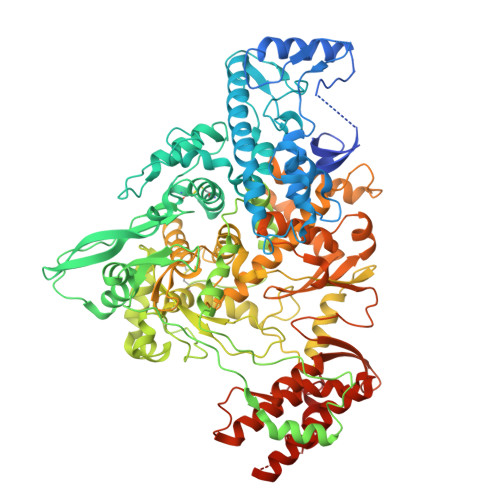
Structural basis of SARS-CoV-2 polymerase inhibition by nonnucleoside inhibitor HeE1-2Tyr.
Kabinger, F., Doze, V., Schmitzova, J., Lidschreiber, M., Dienemann, C., Cramer, P.(2025) Proc Natl Acad Sci U S A 122: e2419854122-e2419854122
- PubMed: 40035759
- DOI: https://doi.org/10.1073/pnas.2419854122
- Primary Citation of Related Structures:
9I81 - PubMed Abstract:
Targeting the RNA-dependent RNA polymerase (RdRp) of SARS-CoV-2 with small molecules is a promising therapeutic strategy against COVID-19, but potent and safe inhibitors are lacking. HeE1-2Tyr, a nonnucleoside inhibitor of Dengue virus RdRp, was also shown to inhibit SARS-CoV-2 RdRp in vitro and to have antiviral activity in cells, but the underlying mechanism remains unclear. Here, we elucidate the molecular mechanism of HeE1-2Tyr-mediated SARS-CoV-2 RdRp inhibition. Biochemical assays confirm that HeE1-2Tyr inhibits RdRp with an IC 50 of 5 µM and show that it competes with RNA binding to RdRp in vitro. Structural analysis using cryo-EM reveals that a stack of three HeE1-2Tyr molecules binds to the RNA binding site of RdRp. The identification of the conserved HeE1-2Tyr binding site and its intriguing inhibition mechanism of three stacked molecules that outcompete RNA may facilitate further development of pan-corona nonnucleoside inhibitors.
- Department of Molecular Biology, Max Planck Institute for Multidisciplinary Sciences, Göttingen 37077, Germany.
Organizational Affiliation: