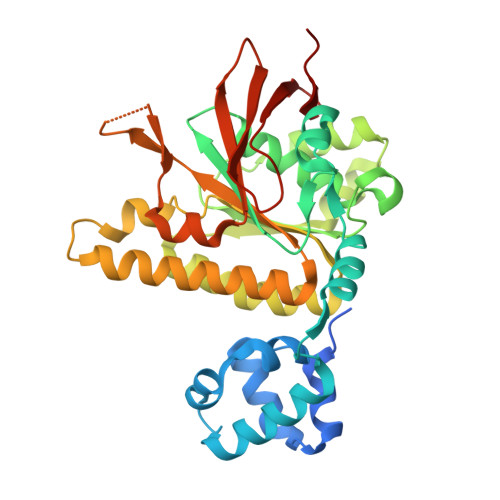
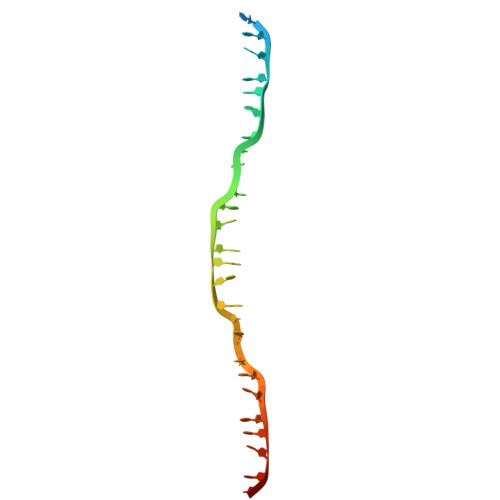
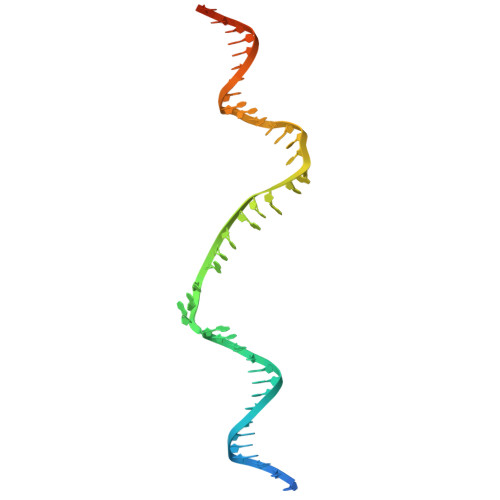
Structural mechanism of strand exchange by the RAD51 filament.
Joudeh, L., Appleby, R.E., Maman, J.D., Pellegrini, L.(2025) Elife 14
- PubMed: 40820711
- DOI: https://doi.org/10.7554/eLife.107114
- Primary Citation of Related Structures:
9I62 - PubMed Abstract:
Homologous recombination (HR) preserves genomic stability by repairing double-strand DNA breaks and ensuring efficient DNA replication. Central to HR is the strand-exchange reaction taking place within the three-stranded synapsis wherein a RAD51 nucleoprotein filament binds to a donor DNA. Here, we present the cryoEM structure of a displacement loop of human RAD51 that captures the synaptic state when the filament has become tightly bound to the donor DNA. The structure elucidates the mechanism of strand exchange by RAD51, including the filament engagement with the donor DNA, the strand invasion and pairing with the complementary sequence of the donor DNA, the capture of the non-complementary strand and the polarity of the strand-exchange reaction. Our findings provide fundamental mechanistic insights into the biochemical reaction of eukaryotic HR.
- Department of Biochemistry, University of Cambridge, Cambridge, United Kingdom.
Organizational Affiliation: