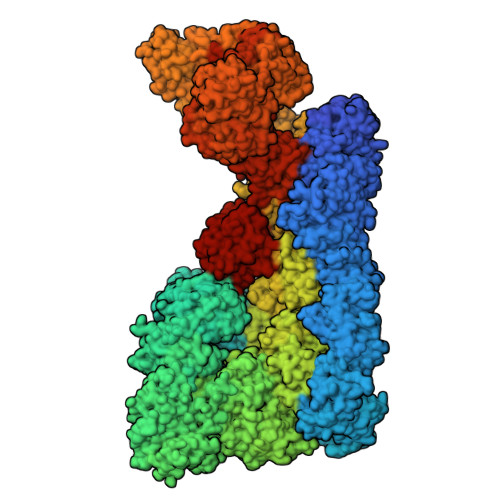
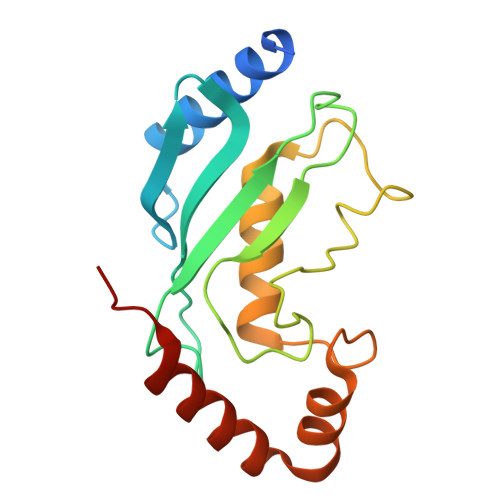
ATP functions as a pathogen-associated molecular pattern to activate the E3 ubiquitin ligase RNF213.
Ahel, J., Balci, A., Faas, V., Grabarczyk, D.B., Harmo, R., Squair, D.R., Zhang, J., Roitinger, E., Lamoliatte, F., Mathur, S., Deszcz, L., Bell, L.E., Lehner, A., Williams, T.L., Sowar, H., Meinhart, A., Wood, N.T., Clausen, T., Virdee, S., Fletcher, A.J.(2025) Nat Commun 16: 4414-4414
- PubMed: 40360510
- DOI: https://doi.org/10.1038/s41467-025-59444-4
- Primary Citation of Related Structures:
9I1I, 9I1J - PubMed Abstract:
The giant E3 ubiquitin ligase RNF213 is a conserved component of mammalian cell-autonomous immunity, limiting the replication of bacteria, viruses and parasites. To understand how RNF213 reacts to these unrelated pathogens, we employ chemical and structural biology to find that ATP binding to its ATPases Associated with diverse cellular Activities (AAA) core activates its E3 function. We develop methodology for proteome-wide E3 activity profiling inside living cells, revealing that RNF213 undergoes a reversible switch in E3 activity in response to cellular ATP abundance. Interferon stimulation of macrophages raises intracellular ATP levels and primes RNF213 E3 activity, while glycolysis inhibition depletes ATP and downregulates E3 activity. These data imply that ATP bears hallmarks of a danger/pathogen associated molecular pattern, coordinating cell-autonomous defence. Furthermore, quantitative labelling of RNF213 with E3-activity probes enabled us to identify the catalytic cysteine required for substrate ubiquitination and obtain a cryo-EM structure of the RNF213-E2-ubiquitin conjugation enzyme transfer intermediate, illuminating an unannotated E2 docking site. Together, our data demonstrate that RNF213 represents a new class of ATP-dependent E3 enzyme, employing distinct catalytic and regulatory mechanisms adapted to its specialised role in the broad defence against intracellular pathogens.
- Research Institute of Molecular Pathology (IMP), Vienna BioCenter, Vienna, Austria.
Organizational Affiliation: