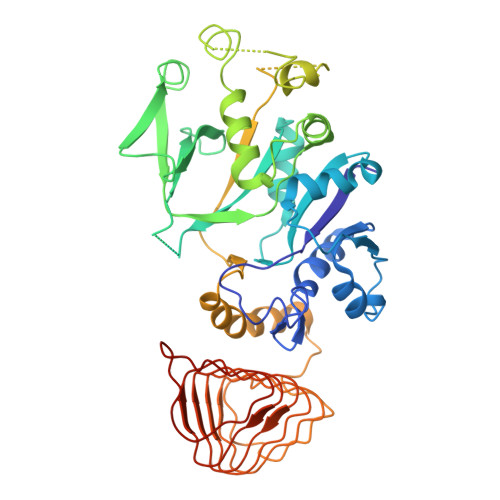
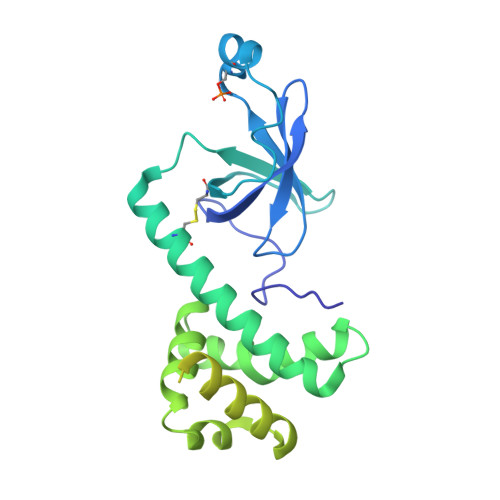
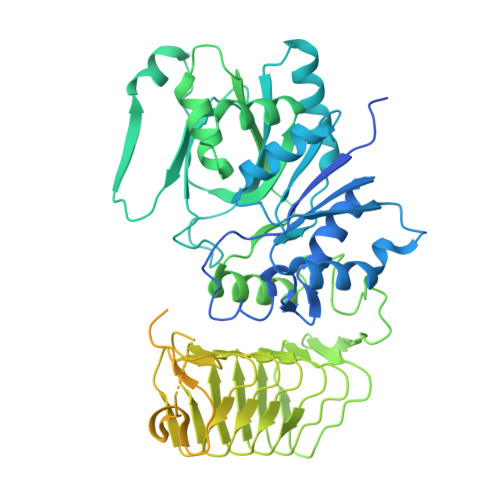
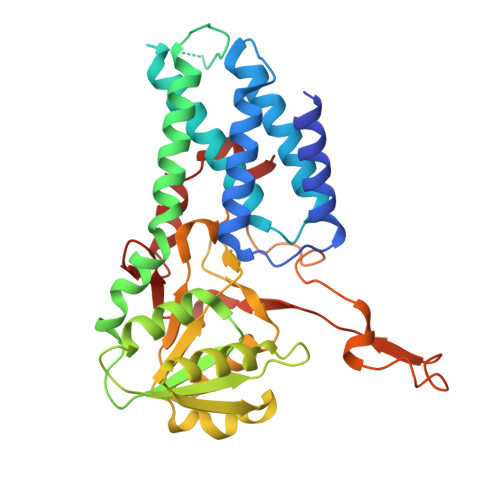
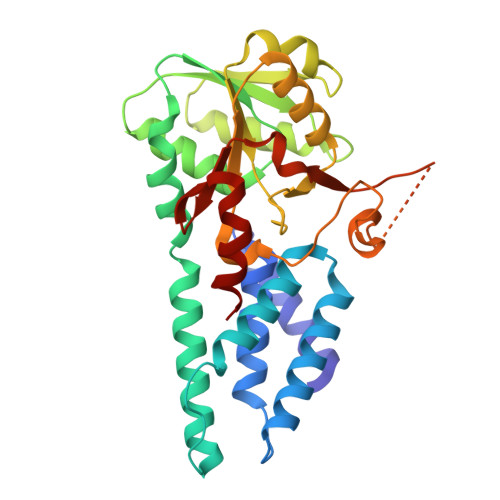
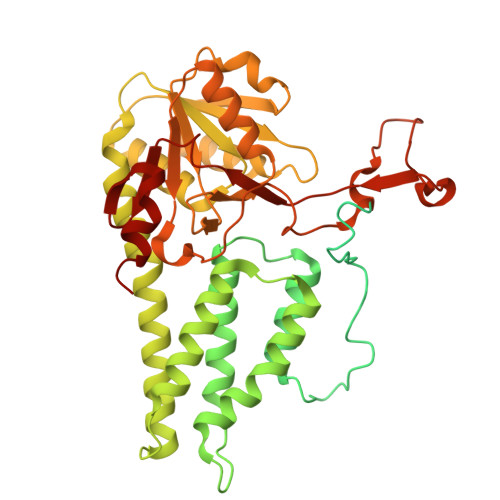
Termination of the integrated stress response.
De Miguel, C., Thorkelsson, S.R., Fatalska, A., Hodgson, G., Wang, C., Bertolotti, A.(2025) Science : eadw5137-eadw5137
- PubMed: 41231936
- DOI: https://doi.org/10.1126/science.adw5137
- Primary Citation of Related Structures:
9HVD, 9HVE, 9HVF - PubMed Abstract:
Stress responses enable cells to detect, adapt to, and survive challenges. The benefit of these signaling pathways depends on their reversibility. The integrated stress response (ISR) is elicited by phosphorylation of translation initiation factor eIF2, which traps and inhibits rate-limiting translation factor eIF2B thereby attenuating translation initiation. Termination of this pathway thus requires relieving eIF2B from P-eIF2 inhibition. Here, we found that eIF2 phosphatase subunits PPP1R15A and PPP1R15B (R15B) bound P-eIF2 in complex with eIF2B. Biochemical investigations guided by cryo-EM structures of native eIF2-eIF2B and P-eIF2-eIF2B complexes bound to R15B demonstrated that R15B enabled dephosphorylation of otherwise dephosphorylation-incompetent P-eIF2 on eIF2B. This sheds light on ISR termination, revealing that R15B rescues eIF2B from P-eIF2 inhibition, thereby safeguarding translation and cell fitness.
- MRC Laboratory of Molecular Biology, Francis Crick Avenue, Cambridge, CB2 0QH, United Kingdom.
Organizational Affiliation: