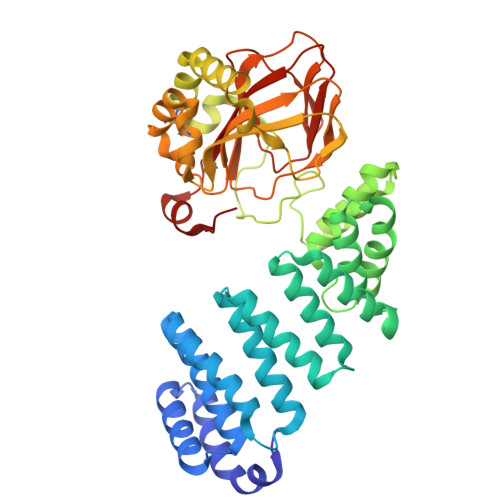
Entry History & Funding Information
Deposition Data
- Released Date: 2025-12-24
Deposition Author(s): de Munnik, M., Rabe, P., Brewitz, L., Brasnett, A., Zhou, T., Schofield, C.J., Kern, J.F.
| Funding Organization | Location | Grant Number |
|---|
| Biotechnology and Biological Sciences Research Council (BBSRC) | United Kingdom | BB/V003291/1 |
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | 1R35GM149528-01 |
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | GM110501 |
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | GM126289 |
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | GM117126 |
- Version 1.0: 2025-12-24
Type: Initial release