Targeted degradation of GSPT1 and NEK7 by a molecular glue prodrug for treatment of HCC.
Glaza, P., Pluta, R., Odrzywol, K.E., Klejnot, M., Wieczorek, M., Cottens, S., Coppen, D., Dobrzanski, P., Drmota, T., Lis-Grzesniak, J., Sniezewska, A., Majkut, J., Mianowska, M., Rozborska, P., Jarmuszkiewicz, M., Kaczanowska, K., Adamska, A., Takagi, T., Sawicka, A., Serwotka-Suszczak, A., Makowska, O., Gajewska, D., Jurczak, K., Leszkowicz, K., Mankiewicz, M., Przytulski, K., Wisniewski, J., Szlachcic, A., Walczak, M.J.(2025) Commun Chem 8: 247-247
- PubMed: 40813917
- DOI: https://doi.org/10.1038/s42004-025-01641-9
- Primary Citation of Related Structures:
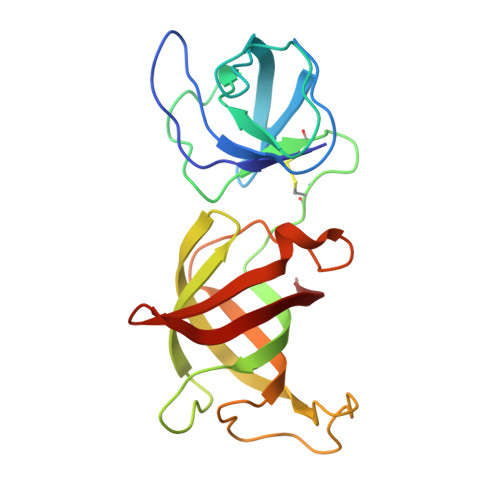
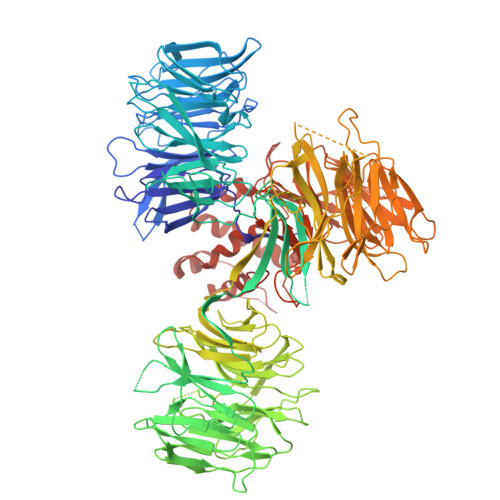
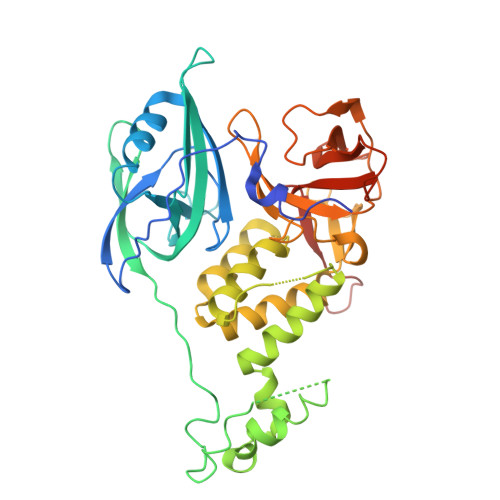
9HNE - PubMed Abstract:
Targeted Protein Degradation (TPD) technology, in the form of CRBN-modulating molecular glues, offers numerous unprecedented therapeutic benefits as evidenced by the success of approved high-value immunomodulatory imide drugs (IMiDs) such as lenalidomide and pomalidomide. Building upon these successes, we employed a small CRBN-focused library of molecular glues in a phenotypic screen against hepatocellular carcinoma (HCC) cell lines. While the original library was primarily designed to target SALL4, we identified additional CRBN substrates, including GSPT1, NEK7, and CK1α, whose degradation potently induced cell death in HCC cell lines. Subsequent lead optimization efforts yielded a compound, ABS-752, which demonstrated superior in vitro and in vivo activity through the potent degradation of GSPT1. Notably, ABS-752 does not form ternary complexes with CRBN and the neosubstrates. Further investigations revealed that ABS-752 is a prodrug activated by the monoamine oxidase, VAP-1, to an aldehyde intermediate and subsequently to the active molecule, ABT-002. VAP-1, which is overexpressed in cirrhotic liver, was identified as the primary monoamine oxidase responsible for the conversion of ABS-752. ABS-752 is currently in clinical trials for the treatment of HCC.
- Captor Therapeutics Inc., Duńska St 11, PL54427, Wrocław, Poland.
Organizational Affiliation: