Unconventional DNA architecture in a dopamine–bound aptamer complex
Chao, E.H.P., Largy, E., Kaiyum, Y.A., Nguyen, M.D., Vialet, B., Dauphin-Ducharme, P., Johnson, P.E., Mackereth, C.D.(2025) bioRxiv
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
(2025) bioRxiv
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| RKEC1 DNA (27-MER) | 27 | synthetic construct | 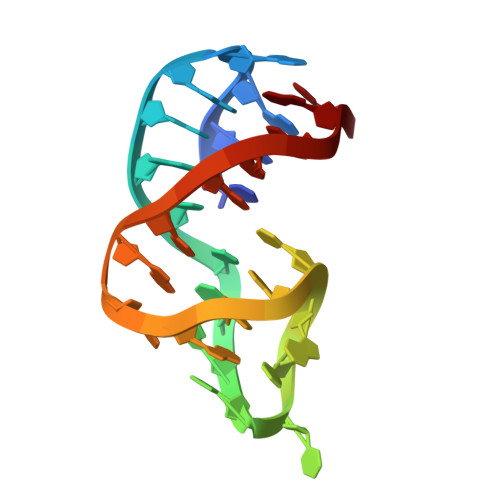 | ||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| LDP (Subject of Investigation/LOI) Query on LDP | B [auth A] | L-DOPAMINE C8 H11 N O2 VYFYYTLLBUKUHU-UHFFFAOYSA-N |  | ||
| Funding Organization | Location | Grant Number |
|---|---|---|
| Other government | Canada | France Canada Research Fund |