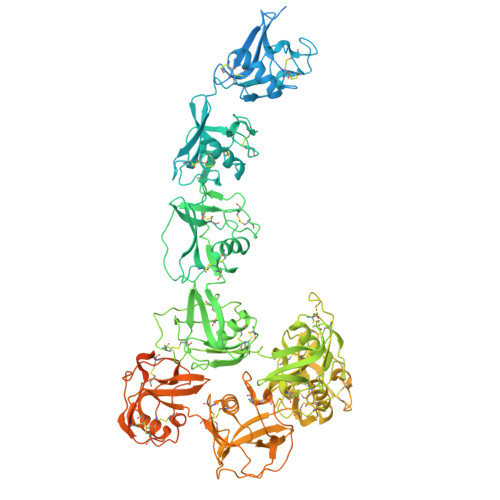
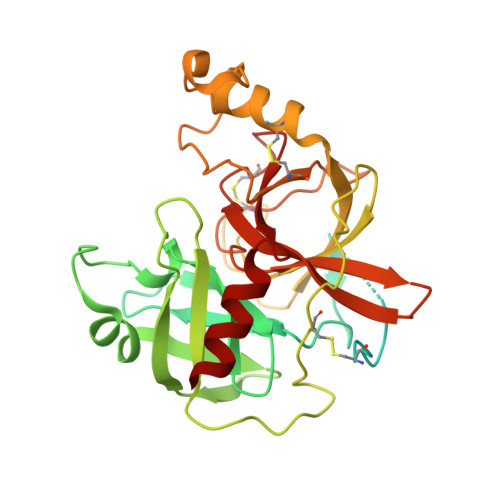
Scavenger receptor CD163 multimerises to allow uptake of diverse ligands.
Zhou, R.X., Higgins, M.K.(2025) Nat Commun 16: 6623-6623
- PubMed: 40681524
- DOI: https://doi.org/10.1038/s41467-025-62054-9
- Primary Citation of Related Structures:
9HEJ, 9HEK, 9HEL - PubMed Abstract:
CD163 is an archetypal scavenger receptor and mediates detoxification of free haemoglobin. Release of haemoglobin from lysed erythrocytes causes oxidative tissue and organ damage. Detoxification involves haemoglobin binding to the abundant serum protein haptoglobin, followed by CD163-mediated uptake of stoichiometrically diverse haptoglobin-haemoglobin complexes into macrophages for degradation. We show that CD163 adopts dimeric and trimeric assemblies due to calcium-mediated interactions within a membrane-associated base. Arms protrude from this base and create a ligand-binding site. Flexibility within the base, coupled with multiple small ligand-binding surfaces on each arm, allow the receptor to mould around its ligands, resulting in promiscuous uptake of ligands with different structures and stoichiometries. Monomeric CD163 lacks this ability to internalise lower-avidity ligands. Arms from adjacent protomers can also self-associate, blocking ligand-binding surfaces in an autoinhibited state. Therefore, through calcium-dependent multimer formation and flexible ligand binding, CD163 scavenges ligands with different structures and avidities, mediating haemoglobin detoxification.
- Department of Biochemistry, University of Oxford, South Parks Road, Oxford, OX1 3QU, UK.
Organizational Affiliation: