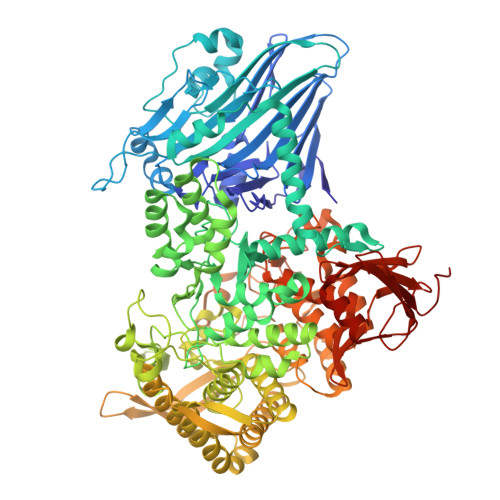
Structural and Functional Dissection of GH161 beta-Glucan Phosphorylases: Molecular Specificities and Dynamics of Catalysis of Dimeric GH-Q Enzymes
Cooper, N.J., Ladeveze, S., Li, A., Shayan, R., Faure, R., Ropartz, D., Terrapon, N., Lombard, V., Henrissat, B., Remaud-Simeon, M., Potocki-Veronese, G., Moulis, C., Cioci, G.(2025) ACS Catal 15: 19312-19327