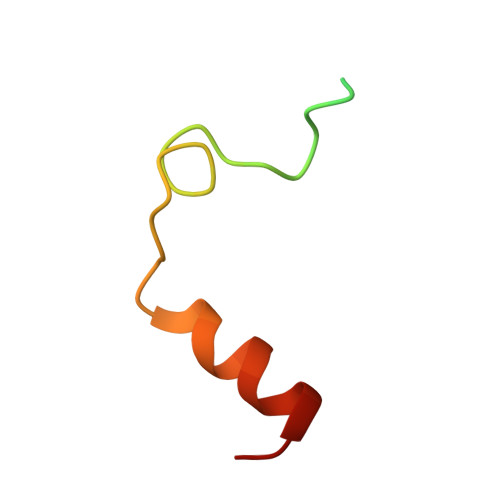
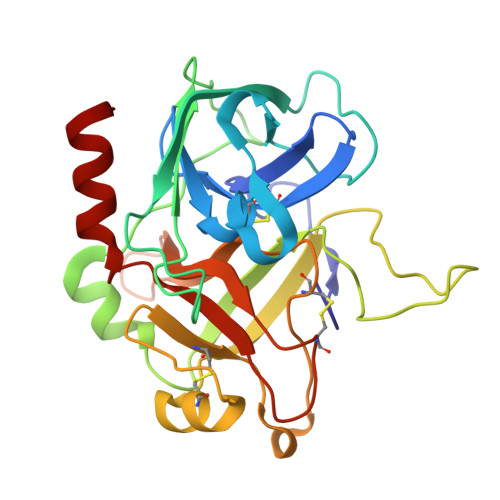
Small molecule ligand development by nanoscale library synthesis and functional screening of crude product
Zsolt, B., Deng, X., Zarda, A., Menoud, G., Will, E., Nielsen, A.R., Farrera-Soler, L., Chinellato, M., Cendron, L., Angelini, A., Heinis, C.To be published.