Cryo-EM structure of DDB1-CRBN in complex with NK7-902 and NEK7
Khoshouei, M., Schroeder, M.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| DNA damage-binding protein 1 | 1,140 | Homo sapiens | Mutation(s): 0 Gene Names: DDB1, XAP1 | 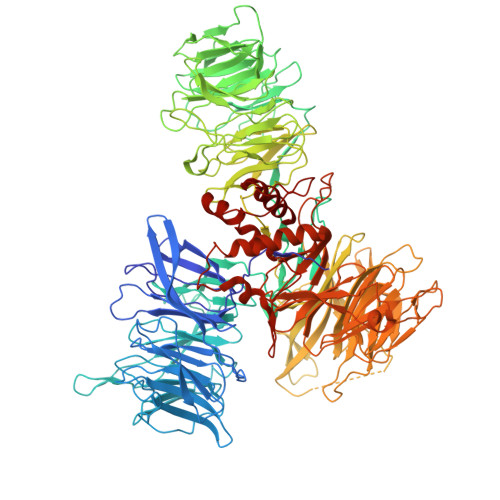 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q16531 (Homo sapiens) Explore Q16531 Go to UniProtKB: Q16531 | |||||
PHAROS: Q16531 GTEx: ENSG00000167986 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q16531 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Serine/threonine-protein kinase Nek7 | B [auth C] | 304 | Homo sapiens | Mutation(s): 0 Gene Names: NEK7 EC: 2.7.11.34 | 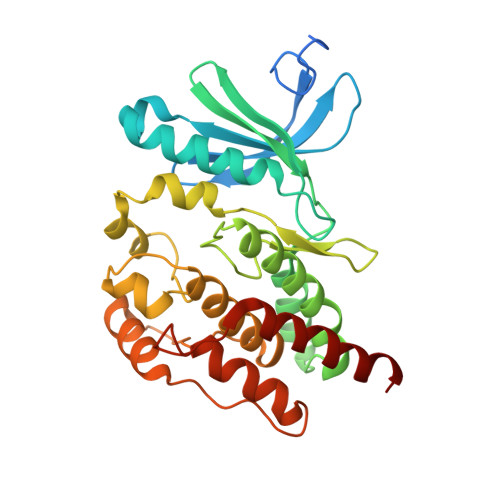 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q8TDX7 (Homo sapiens) Explore Q8TDX7 Go to UniProtKB: Q8TDX7 | |||||
PHAROS: Q8TDX7 GTEx: ENSG00000151414 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q8TDX7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Protein cereblon | C [auth B] | 459 | Homo sapiens | Mutation(s): 0 Gene Names: CRBN, AD-006 | 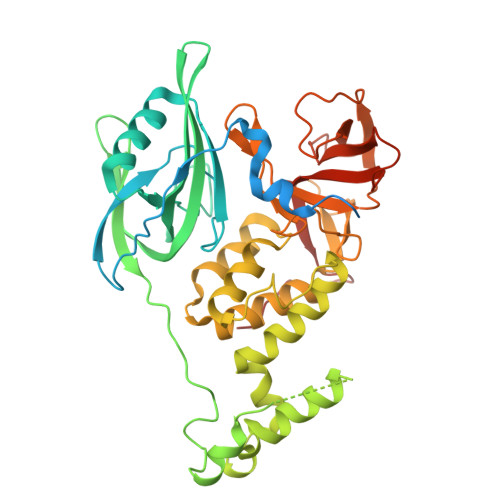 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q96SW2 (Homo sapiens) Explore Q96SW2 Go to UniProtKB: Q96SW2 | |||||
PHAROS: Q96SW2 GTEx: ENSG00000113851 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q96SW2 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| A1ISP (Subject of Investigation/LOI) Query on A1ISP | D [auth B] | 2-[(3S)-2,6-bis(oxidanylidene)piperidin-3-yl]-5-[(1S,2R,5S)-2-(ethylamino)-8-azabicyclo[3.2.1]octan-8-yl]isoindole-1,3-dione C22 H26 N4 O4 QAKJEKTXKXAPBP-LSZYVWPRSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Not funded | -- |