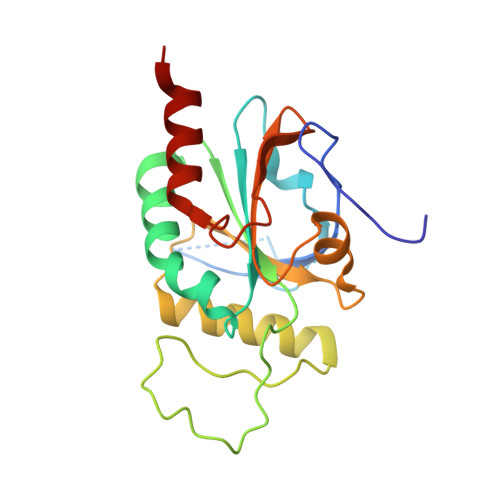
Structural Insights into the Iodothyronine Deiodinase 2 Catalytic Core and Deiodinase Catalysis and Dimerization.
Towell, H., Braun, D., Brol, A., di Fonzo, A., Rijntjes, E., Kohrle, J., Schweizer, U., Steegborn, C.(2024) Biomolecules 14
- PubMed: 39595550
- DOI: https://doi.org/10.3390/biom14111373
- Primary Citation of Related Structures:
9H48 - PubMed Abstract:
Iodothyronine deiodinases (Dio) are selenocysteine-containing membrane enzymes that activate and inactivate the thyroid hormones (TH) through reductive iodide eliminations. The three deiodinase isoforms are homodimers sharing highly conserved amino acid sequences, but they differ in their regioselectivities for the deiodination reaction and regulatory features. We have now solved a crystal structure of the mouse deiodinase 2 (Dio2) catalytic domain. It reveals a high overall similarity to the deiodinase 3 structure, supporting the proposed common mechanism, but also Dio2-specific features, likely mediating its unique properties. Activity studies with an artificially enforced Dio dimer further confirm that dimerization is required for activity and requires both the catalytic core and the enzyme's N-terminus. Cross-linking studies reveal the catalytic core's dimerization interface, providing insights into the architecture of the complete, active Dio homodimer.
- Department of Biochemistry, University of Bayreuth, Universitätsstr. 30, 95447 Bayreuth, Germany.
Organizational Affiliation: