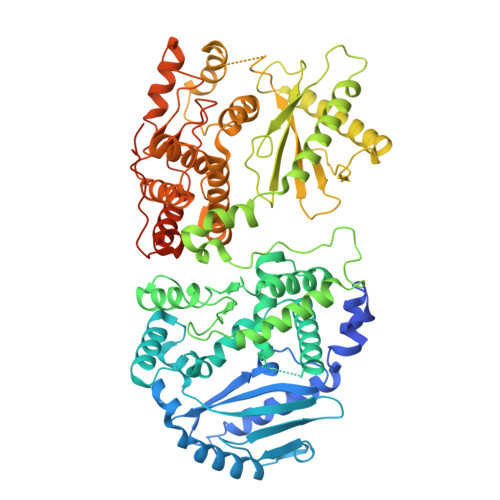
Structural basis for OAS2 regulation and its antiviral function.
Merold, V., Bekere, I., Kretschmer, S., Schnell, A.F., Kmiec, D., Sivarajan, R., Lammens, K., Liu, R., Mergner, J., Teppert, J., Hirschenberger, M., Henrici, A., Hammes, S., Buder, K., Weitz, M., Hackmann, K., Koenig, L.M., Pichlmair, A., Schwierz, N., Sparrer, K.M.J., Lee-Kirsch, M.A., de Oliveira Mann, C.C.(2025) Mol Cell 85: 2176-2193.e13
- PubMed: 40412389
- DOI: https://doi.org/10.1016/j.molcel.2025.05.001
- Primary Citation of Related Structures:
9H1Z - PubMed Abstract:
Oligoadenylate synthetase (OAS) proteins are immune sensors for double-stranded RNA and are critical for restricting viruses. OAS2 comprises two OAS domains, only one of which can synthesize 2'-5'-oligoadenylates for RNase L activation. Existing structures of OAS1 provide a model for enzyme activation, but they do not explain how multiple OAS domains discriminate RNA length. Here, we discover that human OAS2 exists in an auto-inhibited state as a zinc-mediated dimer and present a mechanism for RNA length discrimination: the catalytically deficient domain acts as a molecular ruler that prevents autoreactivity to short RNAs. We demonstrate that dimerization and myristoylation localize OAS2 to Golgi membranes and that this is required for OAS2 activation and the restriction of viruses that exploit the endomembrane system for replication, e.g., coronaviruses. Finally, our results highlight the non-redundant role of OAS proteins and emphasize the clinical relevance of OAS2 by identifying a patient with a loss-of-function mutation associated with autoimmune disease.
- Department of Bioscience, TUM School of Natural Sciences, Technical University of Munich, Garching 85748, Germany.
Organizational Affiliation: