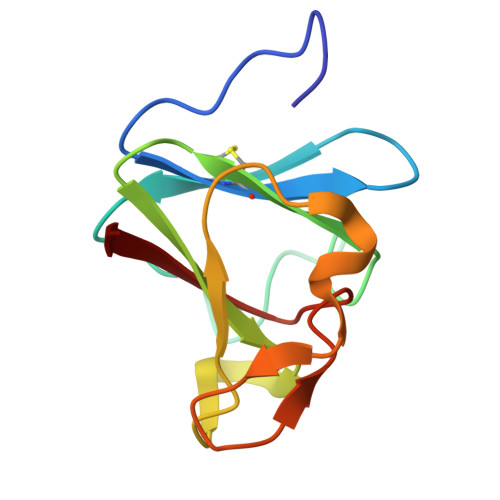
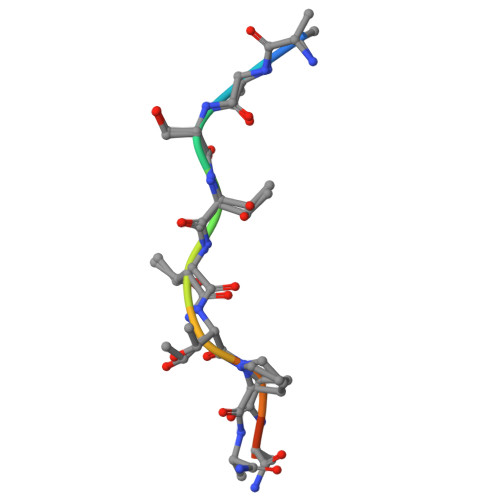
Microbial binding module employs sophisticated clustered saccharide patches to selectively adhere to mucins.
Jaroentomeechai, T., Veloz, B., Soares, C.O., Goerdeler, F., Grosso, A.S., Bull, C., Miller, R.L., Furukawa, S., Gines-Alcober, I., Taleb, V., Merino, P., Ghirardello, M., Companon, I., Coelho, H., Dias, J.S., Vincentelli, R., Henrissat, B., Joshi, H., Clausen, H., Corzana, F., Marcelo, F., Hurtado-Guerrero, R., Narimatsu, Y.(2025) Nat Commun 16: 9058-9058
- PubMed: 41083434
- DOI: https://doi.org/10.1038/s41467-025-63756-w
- Primary Citation of Related Structures:
9GRF, 9GRJ, 9GSM - PubMed Abstract:
The mucus lining wet body surfaces forms the interphase and barrier for the microbiota and resident microbiomes. Large mucin proteins densely decorated with O-glycans make up the mucus lining to entrap, feed and shape the microbiota, and repress biofilm formation and virulence. How mucins exert these effects is poorly understood and critical is how the microbiota recognize, sense, and break down mucins. Here, we provide structural molecular evidence that a small mucin-binding module designated X409 recognizes clustered saccharide patches comprised of rows of inner monosaccharides in adjacent O-glycans. These patches are unique to mucins and binding to these provides an elegant mechanism to retain adherence to mucins despite trimming of O-glycans during microbial scavenging of monosaccharides from mucins. Realization of clustered saccharide patch-binding motifs provides a hitherto overlooked scenario of contextual glycan epitopes and impetus for discovery of new classes of glycan-binding proteins.
- Copenhagen Center for Glycomics, Departments of Cellular and Molecular Medicine, Faculty of Health Sciences, University of Copenhagen, Copenhagen, Denmark.
Organizational Affiliation: