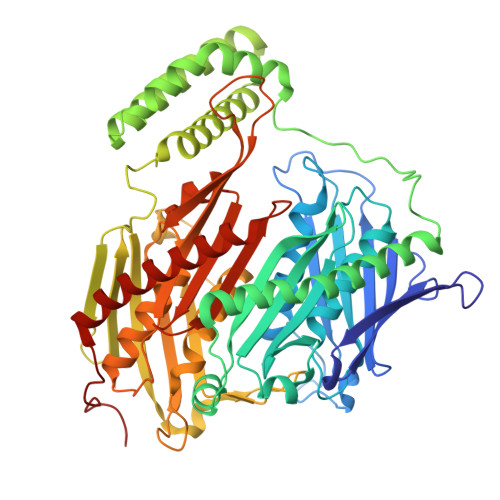
Selective inhibition of Mycobacterium tuberculosis GpsI unveils a novel strategy to target the RNA metabolism.
Griesser, T., Wang, R., Angona, I.P., Rogenmoser, J., Obrist, J., Schneider, G., Sander, P.(2025) Nucleic Acids Res 53
- PubMed: 40530695
- DOI: https://doi.org/10.1093/nar/gkaf529
- Primary Citation of Related Structures:
9GMS, 9GMT - PubMed Abstract:
Polyribonucleotide nucleotidyl-transferases (PNPases) play a critical role in the degradation of mRNA. The mycobacterial PNPase, guanosine penta-phosphate synthase I (GpsI), is an essential enzyme in Mycobacterium tuberculosis (Mtb), collaborating with endoribonucleases and helicases to process RNA. In this study, we identify GpsI as a novel and underexplored drug target. The inhibitor 1-(4'-(2-phenyl-5-(trifluoromethyl) oxazole-4-carboxamido)-[1,1'-biphenyl]-4-caroxamido) cyclopentane-1-carboxylic acid (X1), discovered through a whole-cell screening, specifically inhibits GpsI activity in biochemical assays. Biochemical and physiological analyses of engineered GpsI variants and recombinant Mycobacterium smegmatis pinpoint amino acids 328 and 527 as critical residues for the selective activity of X1 against Mtb complex. High-resolution cryo-electron microscopy analysis of the ternary GpsI-X1-poly(A) complex elucidates the drug-binding pocket, providing insight into its mechanism of action. This study introduces a potent inhibitor targeting the underexplored Mtb-GpsI and offers a molecular explanation for its selective specificity.
- Institute of Medical Microbiology, University of Zurich, 8006 Zurich, Switzerland.
Organizational Affiliation: