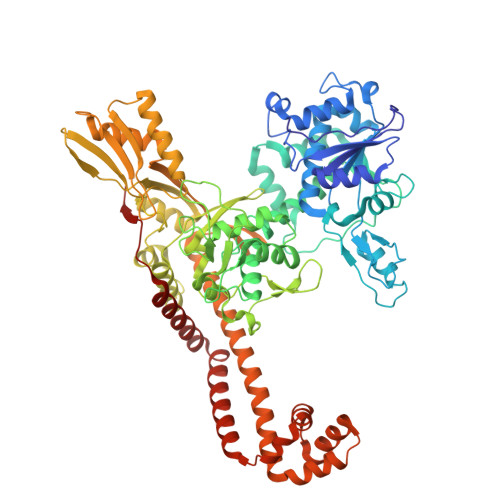
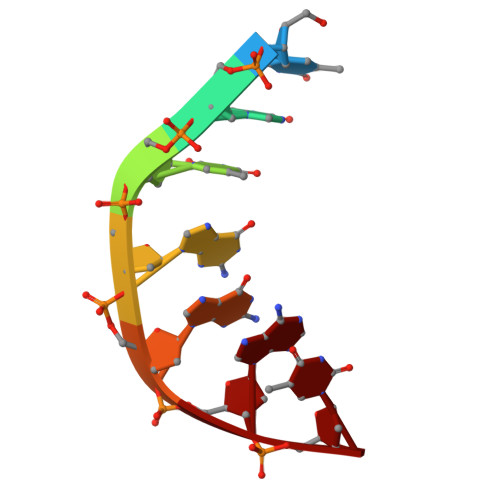
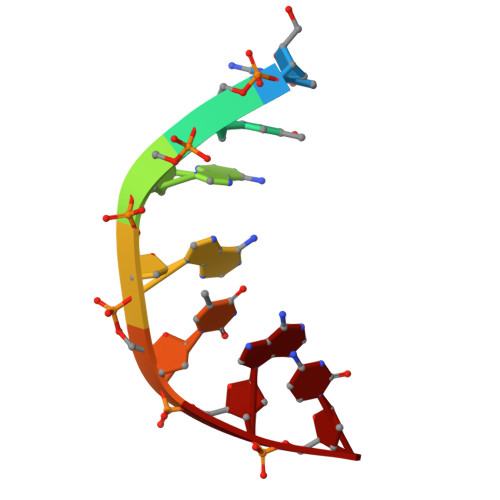
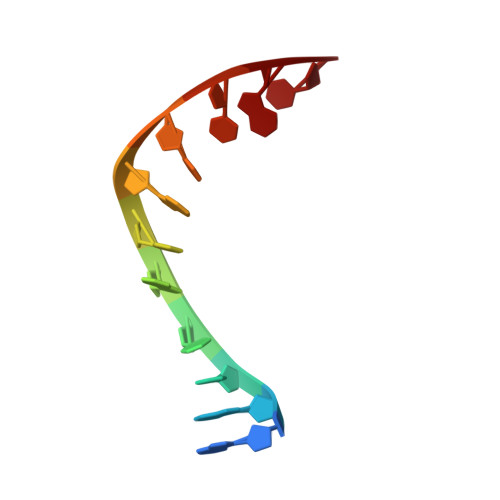
Experimental localization of metal-binding sites reveals the role of metal ions in type II DNA topoisomerases.
Wang, B., Najmudin, S., Pan, X.S., Mykhaylyk, V., Orr, C., Wagner, A., Govada, L., Chayen, N.E., Fisher, L.M., Sanderson, M.R.(2024) Proc Natl Acad Sci U S A 121: e2413357121-e2413357121
- PubMed: 39361644
- DOI: https://doi.org/10.1073/pnas.2413357121
- Primary Citation of Related Structures:
9GEF - PubMed Abstract:
Metal ions have important roles in supporting the catalytic activity of DNA-regulating enzymes such as topoisomerases (topos). Bacterial type II topos, gyrases and topo IV, are primary drug targets for fluoroquinolones, a class of clinically relevant antibacterials requiring metal ions for efficient drug binding. While the presence of metal ions in topos has been elucidated in biochemical studies, accurate location and assignment of metal ions in structural studies have historically posed significant challenges. Recent advances in X-ray crystallography address these limitations by extending the experimental capabilities into the long-wavelength range, exploiting the anomalous contrast from light elements of biological relevance. This breakthrough enables us to confirm experimentally the locations of Mg 2+ in the fluoroquinolone-stabilized Streptococcus pneumoniae topo IV complex. Moreover, we can unambiguously identify the presence of K + and Cl - ions in the complex with one pair of K + ions functioning as an additional intersubunit bridge. Overall, our data extend current knowledge on the functional and structural roles of metal ions in type II topos.
- Department of Metabolism, Digestion and Reproduction, Faculty of Medicine, Imperial College London, London W12 0NN, United Kingdom.
Organizational Affiliation: