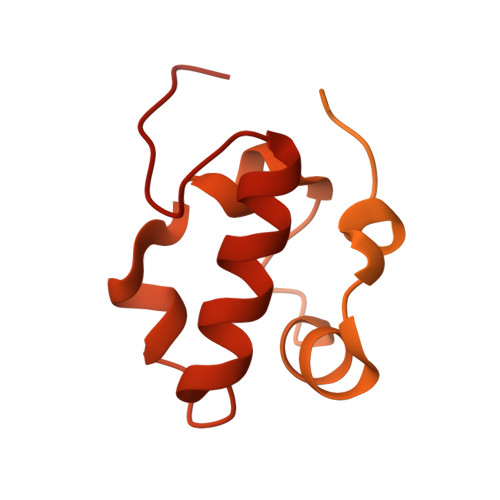
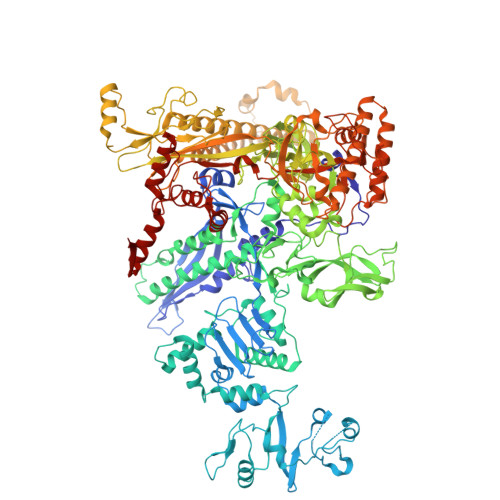
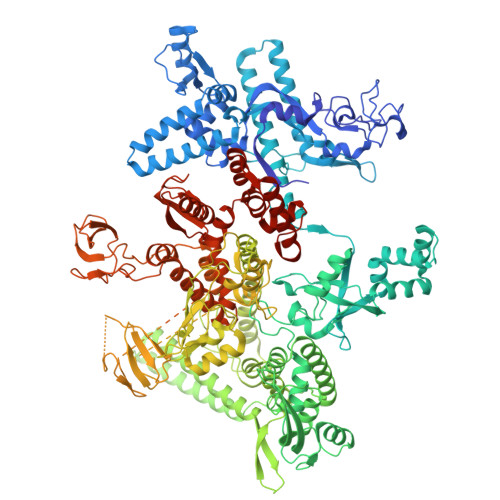
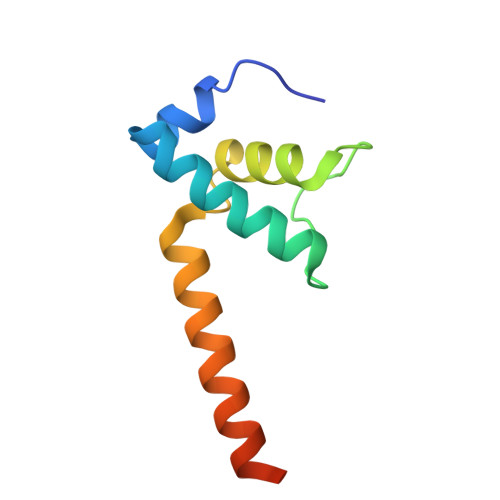
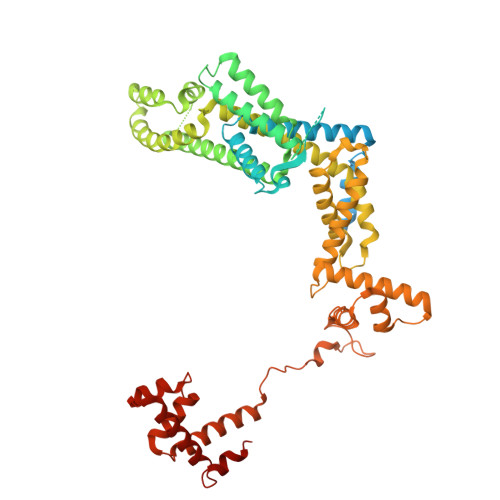
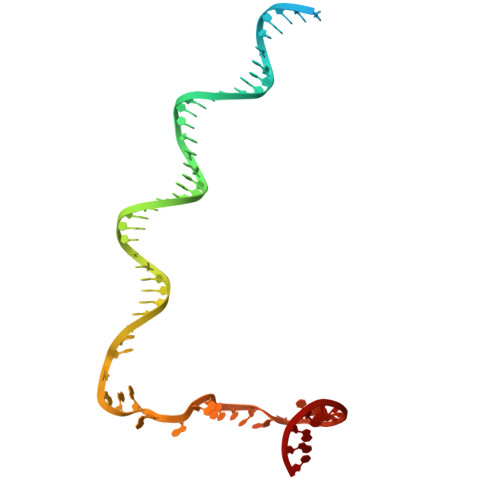
Structures of Vibrio cholerae transcription complexes reveal how ToxR and TcpP recruit the RNA polymerase and activate virulence genes.
Alcaide-Jimenez, A., Canals, A., Baudin, F., Machon, C., Fabrega-Ferrer, M., Bantysh, O., Perez-Luque, R., Murciano, B., Mohammad, A.A., Rowse, M.J., Ferracciolo, J.M., Krukonis, E.S., Muller, C.W., Coll, M.(2026) Sci Adv 12: eadx9680-eadx9680
- PubMed: 41533787
- DOI: https://doi.org/10.1126/sciadv.adx9680
- Primary Citation of Related Structures:
9GDO, 9GDP, 9GDQ, 9GDR, 9GDS - PubMed Abstract:
Activation of virulence in Vibrio cholerae , the etiological agent of cholera disease, is mediated by two transmembrane one-component signal-transduction proteins, ToxR and TcpP, which are also transcription factors. Using cryo-electron microscopy, we have solved five structures of the ompU and toxT transcription activation complexes, including the RNA polymerase (RNAP) holoenzyme, promoter DNAs, transcribed RNA, and their corresponding transcription factors, ToxR or TcpP and ToxR-TcpP, respectively. Activation is achieved through the interaction of ToxR or TcpP with the α-C-terminal repeat domain of RNAP where a single residue of the activator, a phenylalanine, appears to be the most critical contact, as confirmed by mutagenesis. No interactions of the transcription factors were observed with other subunits of the RNAP, i.e., the σ subunit as it occurs in the structurally related PhoB family of two-component transcription factors. The structures, and their comparison with our previously solved DNA promoter-ToxR x-ray structures, unveil the molecular mechanism of cholera virulence gene activation.
- Institute for Research in Biomedicine (IRB Barcelona), Barcelona Institute of Science and Technology (BIST), Baldiri Reixac 10-12, 08028 Barcelona, Spain.
Organizational Affiliation: