Inactive PSII dimer from the Peak4 PSII pool
Zhao, Z., Vercellino, I., Nixon, P.J., Sazanov, L.A.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem II protein D1 1 | A, Q [auth a] | 360 | Thermosynechococcus vestitus BP-1 | Mutation(s): 0 EC: 1.10.3.9 | 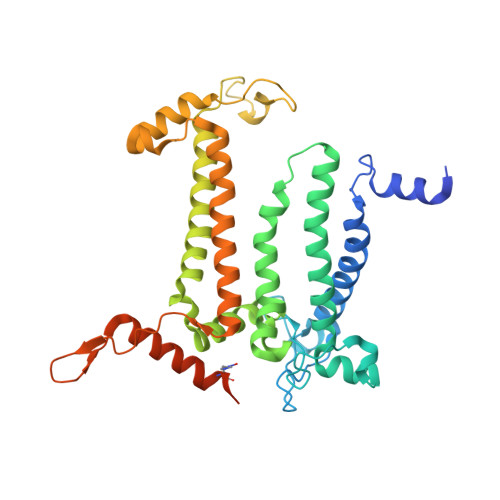 |
UniProt | |||||
Find proteins for P0A444 (Thermosynechococcus vestitus (strain NIES-2133 / IAM M-273 / BP-1)) Explore P0A444 Go to UniProtKB: P0A444 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0A444 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem II CP47 reaction center protein | B, R [auth b] | 510 | Thermosynechococcus vestitus BP-1 | Mutation(s): 0 | 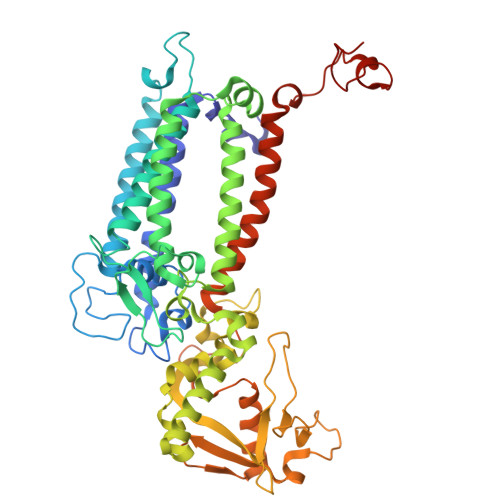 |
UniProt | |||||
Find proteins for Q8DIQ1 (Thermosynechococcus vestitus (strain NIES-2133 / IAM M-273 / BP-1)) Explore Q8DIQ1 Go to UniProtKB: Q8DIQ1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q8DIQ1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem II CP43 reaction center protein | C, S [auth c] | 461 | Thermosynechococcus vestitus BP-1 | Mutation(s): 0 | 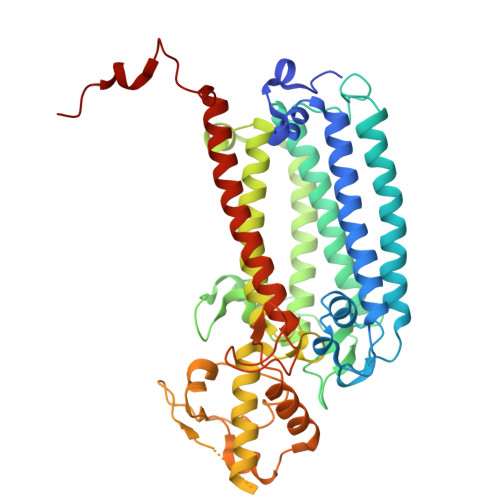 |
UniProt | |||||
Find proteins for Q8DIF8 (Thermosynechococcus vestitus (strain NIES-2133 / IAM M-273 / BP-1)) Explore Q8DIF8 Go to UniProtKB: Q8DIF8 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q8DIF8 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem II D2 protein | D, T [auth d] | 352 | Thermosynechococcus vestitus BP-1 | Mutation(s): 0 EC: 1.10.3.9 | 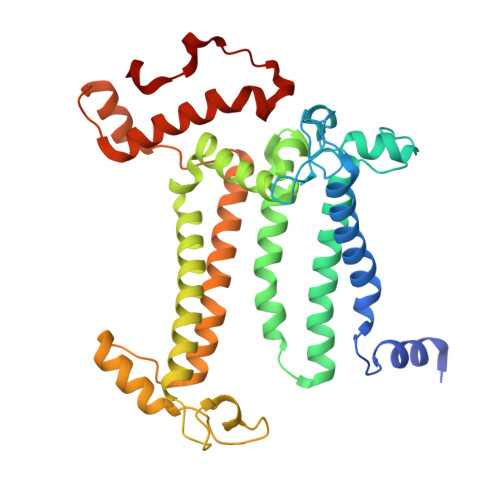 |
UniProt | |||||
Find proteins for Q8CM25 (Thermosynechococcus vestitus (strain NIES-2133 / IAM M-273 / BP-1)) Explore Q8CM25 Go to UniProtKB: Q8CM25 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q8CM25 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cytochrome b559 subunit alpha | E, U [auth e] | 84 | Thermosynechococcus vestitus BP-1 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q8DIP0 (Thermosynechococcus vestitus (strain NIES-2133 / IAM M-273 / BP-1)) Explore Q8DIP0 Go to UniProtKB: Q8DIP0 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q8DIP0 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 6 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cytochrome b559 subunit beta | F, V [auth f] | 45 | Thermosynechococcus vestitus BP-1 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q8DIN9 (Thermosynechococcus vestitus (strain NIES-2133 / IAM M-273 / BP-1)) Explore Q8DIN9 Go to UniProtKB: Q8DIN9 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q8DIN9 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 7 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem II reaction center protein H | G [auth H], W [auth h] | 66 | Thermosynechococcus vestitus BP-1 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q8DJ43 (Thermosynechococcus vestitus (strain NIES-2133 / IAM M-273 / BP-1)) Explore Q8DJ43 Go to UniProtKB: Q8DJ43 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q8DJ43 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 8 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem II reaction center protein I | H [auth I], X [auth i] | 38 | Thermosynechococcus vestitus BP-1 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q8DJZ6 (Thermosynechococcus vestitus (strain NIES-2133 / IAM M-273 / BP-1)) Explore Q8DJZ6 Go to UniProtKB: Q8DJZ6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q8DJZ6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 9 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem II reaction center protein J | I [auth J], Y [auth j] | 40 | Thermosynechococcus vestitus BP-1 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P59087 (Thermosynechococcus vestitus (strain NIES-2133 / IAM M-273 / BP-1)) Explore P59087 Go to UniProtKB: P59087 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P59087 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 10 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem II reaction center protein K | J [auth K], Z [auth k] | 46 | Thermosynechococcus vestitus BP-1 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q9F1K9 (Thermosynechococcus vestitus (strain NIES-2133 / IAM M-273 / BP-1)) Explore Q9F1K9 Go to UniProtKB: Q9F1K9 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9F1K9 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 11 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem II reaction center protein L | AA [auth l], K [auth L] | 37 | Thermosynechococcus vestitus BP-1 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q8DIN8 (Thermosynechococcus vestitus (strain NIES-2133 / IAM M-273 / BP-1)) Explore Q8DIN8 Go to UniProtKB: Q8DIN8 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q8DIN8 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 12 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem II reaction center protein M | BA [auth m], L [auth M] | 36 | Thermosynechococcus vestitus BP-1 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q8DHA7 (Thermosynechococcus vestitus (strain NIES-2133 / IAM M-273 / BP-1)) Explore Q8DHA7 Go to UniProtKB: Q8DHA7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q8DHA7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 13 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem II reaction center protein T | CA [auth t], M [auth T] | 32 | Thermosynechococcus vestitus BP-1 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q8DIQ0 (Thermosynechococcus vestitus (strain NIES-2133 / IAM M-273 / BP-1)) Explore Q8DIQ0 Go to UniProtKB: Q8DIQ0 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q8DIQ0 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 14 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem II reaction center X protein | DA [auth x], N [auth X] | 41 | Thermosynechococcus vestitus BP-1 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q9F1R6 (Thermosynechococcus vestitus (strain NIES-2133 / IAM M-273 / BP-1)) Explore Q9F1R6 Go to UniProtKB: Q9F1R6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9F1R6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 15 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem II reaction center protein Ycf12 | EA [auth y], O [auth Y] | 46 | Thermosynechococcus vestitus BP-1 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q8DJI1 (Thermosynechococcus vestitus (strain NIES-2133 / IAM M-273 / BP-1)) Explore Q8DJI1 Go to UniProtKB: Q8DJI1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q8DJI1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 16 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem II reaction center protein Z | FA [auth z], P [auth Z] | 62 | Thermosynechococcus vestitus BP-1 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q8DHJ2 (Thermosynechococcus vestitus (strain NIES-2133 / IAM M-273 / BP-1)) Explore Q8DHJ2 Go to UniProtKB: Q8DHJ2 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q8DHJ2 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 16 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| DGD Query on DGD | JD [auth D] LH [auth d] PD [auth H] QC [auth C] QH [auth h] | DIGALACTOSYL DIACYL GLYCEROL (DGDG) C51 H96 O15 LDQFLSUQYHBXSX-HXXRYREZSA-N |  | ||
| CLA Query on CLA | AB [auth B] AF [auth b] BB [auth B] BF [auth b] BH [auth d] | CHLOROPHYLL A C55 H72 Mg N4 O5 ATNHDLDRLWWWCB-AENOIHSZSA-M |  | ||
| PHO Query on PHO | LA [auth A], OE [auth a], VC [auth D], ZG [auth d] | PHEOPHYTIN A C55 H74 N4 O5 CQIKWXUXPNUNDV-RCBXBCQGSA-N |  | ||
| SQD Query on SQD | BE [auth L] BI [auth l] MD [auth F] OA [auth A] OH [auth f] | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL C41 H78 O12 S RVUUQPKXGDTQPG-JUDHQOGESA-N |  | ||
| LMG Query on LMG | AC [auth C] EG [auth c] HH [auth d] ID [auth D] QB [auth B] | 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE C45 H86 O10 DCLTVZLYPPIIID-CVELTQQQSA-N |  | ||
| PL9 Query on PL9 | EH [auth d], FD [auth D], PA [auth A], SE [auth a] | 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE C53 H80 O2 FKUYMLZIRPABFK-UHFFFAOYSA-N |  | ||
| LHG Query on LHG | AE [auth L] AI [auth l] FH [auth d] GD [auth D] GH [auth d] | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE C38 H75 O10 P BIABMEZBCHDPBV-MPQUPPDSSA-N |  | ||
| HEM Query on HEM | LD [auth F], NH [auth f] | PROTOPORPHYRIN IX CONTAINING FE C34 H32 Fe N4 O4 KABFMIBPWCXCRK-RGGAHWMASA-L |  | ||
| RRX Query on RRX | HI [auth x], OD [auth H] | (3R)-beta,beta-caroten-3-ol C40 H56 O DMASLKHVQRHNES-FKKUPVFPSA-N |  | ||
| BCR Query on BCR | DH [auth d] ED [auth D] GE [auth Y] II [auth y] NA [auth A] | BETA-CAROTENE C40 H56 OENHQHLEOONYIE-JLTXGRSLSA-N |  | ||
| LMT Query on LMT | BC [auth C] CE [auth M] CI [auth m] DE [auth M] DI [auth m] | DODECYL-BETA-D-MALTOSIDE C24 H46 O11 NLEBIOOXCVAHBD-QKMCSOCLSA-N |  | ||
| LFA Query on LFA | AD [auth D] AG [auth b] BD [auth D] DG [auth b] EE [auth M] | EICOSANE C20 H42 CBFCDTFDPHXCNY-UHFFFAOYSA-N |  | ||
| PLM Query on PLM | BG [auth b] CG [auth b] IE [auth a] KD [auth E] MH [auth e] | PALMITIC ACID C16 H32 O2 IPCSVZSSVZVIGE-UHFFFAOYSA-N |  | ||
| BCT Query on BCT | AH [auth d], VA [auth A] | BICARBONATE ION C H O3 BVKZGUZCCUSVTD-UHFFFAOYSA-M |  | ||
| FE2 Query on FE2 | GA [auth A], JE [auth a] | FE (II) ION Fe CWYNVVGOOAEACU-UHFFFAOYSA-N |  | ||
| CL Query on CL | HA [auth A], KE [auth a] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| Modified Residues 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| HSK Query on HSK | A, Q [auth a] | L-PEPTIDE LINKING | C6 H9 N3 O3 |  | HIS |
| FME Query on FME | H [auth I], X [auth i] | L-PEPTIDE LINKING | C6 H11 N O3 S |  | MET |
| Task | Software Package | Version |
|---|---|---|
| RECONSTRUCTION | RELION | 4.0.0 |
| MODEL REFINEMENT | PHENIX | dev_4933+SVN |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Biotechnology and Biological Sciences Research Council (BBSRC) | United Kingdom | BB/I00937X/1 |
| Biotechnology and Biological Sciences Research Council (BBSRC) | United Kingdom | BB/V002007/1 |